| Customer Name | ####### | Customer Institution | ####### | Customer Email | ####### | Project ID | ####### |
|---|---|
| Seller Name | Kyle Navel | Seller Email | knavel@lcsciences.com |
Your reliable partner in genomics, transcriptomics and proteomics
N6-Methyladenosine (m6A) is an abundant modification in mRNA and is found within some viruses, and most eukaryotes including mammals, insects, plants and yeast. It is also found in tRNA, rRNA, and small nuclear RNA (snRNA) as well as several long non-coding RNA.
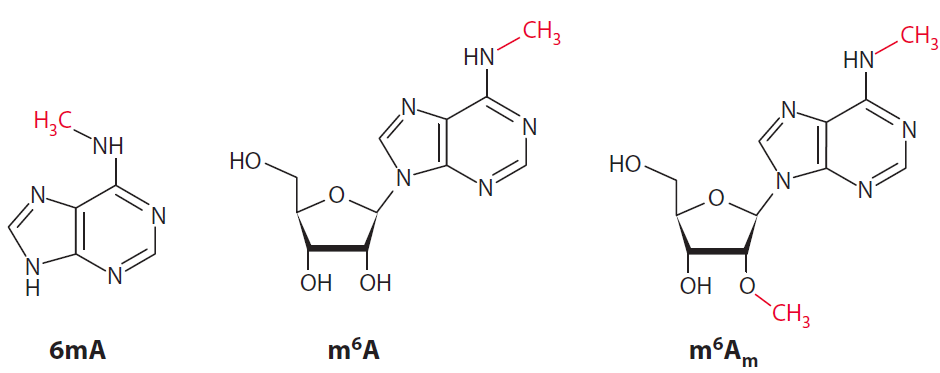
m6A and m6Am are the two 6mA (6-methyladenine)-containing nucleotides in mRNA. 6mA is found in two distinct epitranscriptomic modifications: m6A (N6-methyladenosine) and m6Am (N6,2'-Odimethyladenosine). The methyl groups that are added enzymatically to alter the function of the nucleotide are shown in red. m6A is found in 5'UTRs, coding sequences, and 3'UTRs. m6Am is found in one place in mRNAs, the first encoded nucleotide, which is adjacent to the m7G (N7-methylguanosine) cap.
The methylation of adenosine is directed by a large m6A methyltransferase complex containing METTL3 as the SAM-binding sub-unit. In vitro, this methyltransferase complex preferentially methylates RNA oligonucleotides containing RRACH motif and a similar preference was identified in vivo in mapped m6A sites in Rous sarcoma virus genomic RNA and in bovine prolactin mRNA. Wilms' tumor 1-associating protein (WTAP) may function to regulate recruitment of the m6A methyltransferase complex to mRNA targets. More recent studies have characterized other key components of the m6A methyltransferase complex in mammals, including METTL14, KIAA1429(Vir), ZFP217, RMB15, RBM15b, HAKAI(CBLL1), ZC3H13 and other proteins. Following a 2010 speculation of m6A in mRNA being dynamic and reversible, the discovery of the first m6A demethylase, fat mass and obesity-associated protein (FTO) in 2011 confirmed this hypothesis and revitalized the interests in the study of m6A by He, Chuan in The University of Chicago. A second m6A demethylase alkB homolog 5 (ALKBH5) was later discovered as well.
The biological functions of m6A are mediated through a group of RNA binding proteins that specifically recognize the methylated adenosine on RNA. These binding proteins are named m6A readers. The YT521-B homology (YTH) domain family of proteins (YTHDF1, YTHDF2, YTHDF3, YTHDC1 and YTHDC2) have been characterized as direct m6A readers and have a conserved m6A-binding pocket. Insulin-like growth factor-2 mRNA-binding proteins 1, 2, and 3 (IGF2BP1–3) are reported as a novel class of m6A readers. IGF2BPs use K homology (KH) domains to selectively recognize m6A-containing RNAs and promote their translation and stability. These m6A readers, together with m6A methyltransferases (writers) and demethylases (erasers), establish a complex mechanism of m6A regulation in which writers and erasers determine the distributions of m6A on RNA, whereas readers mediate m6A-dependent functions. m6A has also been shown to mediate a structural switch termed m6A switch.
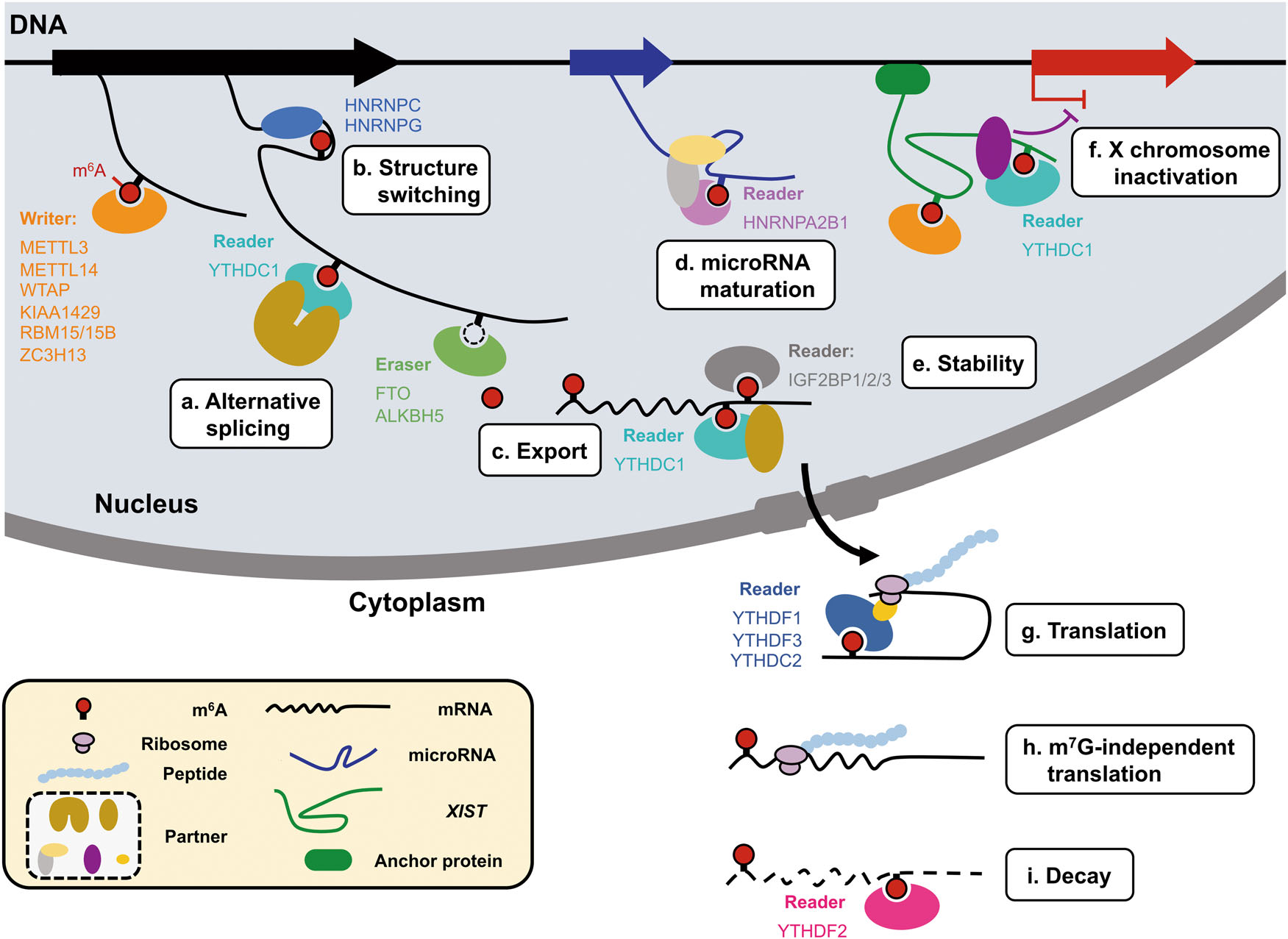
A review named Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism published on Cell Research described the current understanding of the m6A modification, particularly the functions of its writers, erasers, readers in RNA metabolism, with an emphasis on its role in regulating the isoform dosage of mRNAs.
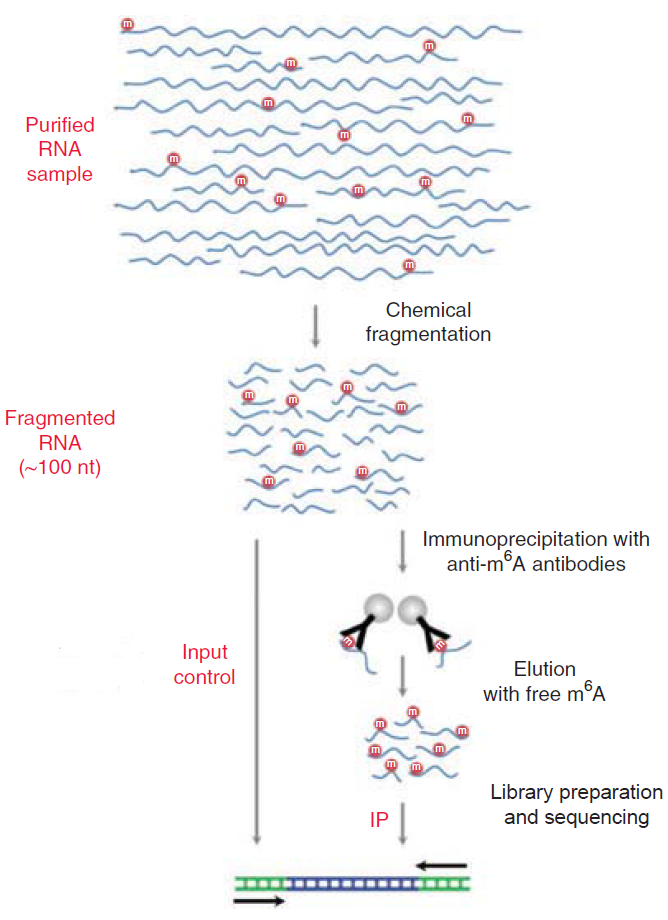
Total RNA was extracted using Trizol reagent (Invitrogen, CA, USA) following the manufacturer's procedure. The total RNA quality and quantity were analysis of Bioanalyzer 2100 and RNA 6000 Nano LabChip Kit (Agilent, CA, USA) with RIN number >7.0. Approximately more than 50 ug of total RNA was subjected to isolate Poly (A) mRNA with poly-T oligo attached magnetic beads (Invitrogen). Following purification, the poly(A) mRNA fractions is fragmented into ~100-nt-long oligonucleotides using divalent cations under elevated temperature. Then the cleaved RNA fragments were subjected to incubated for 2h at 4℃ with m6A-specific antibody (No. 202003, Synaptic Systems, Germany) in IP buffer (50 mM Tris-HCl, 750 mM NaCl and 0.5% Igepal CA-630) supplemented with BSA (0.5 μg μl−1). The mixture was then incubated with protein-A beads and eluted with elution buffer (1 × IP buffer and 6.7mM m6A). Eluted RNA was precipitated by 75% ethanol. Eluted m6A-containing fragments (IP) and untreated input control fragments are converted to final cDNA library in accordance with a strand-specific library preparation by dUTP method. The average insert size for the paired-end libraries was ~100±50 bp. And then we performed the paired-end 2×150bp sequencing on an Illumina Novaseq™ 6000 platform at the LC-BIO Bio-tech ltd (Hangzhou, China) following the vendor's recommended protocol.
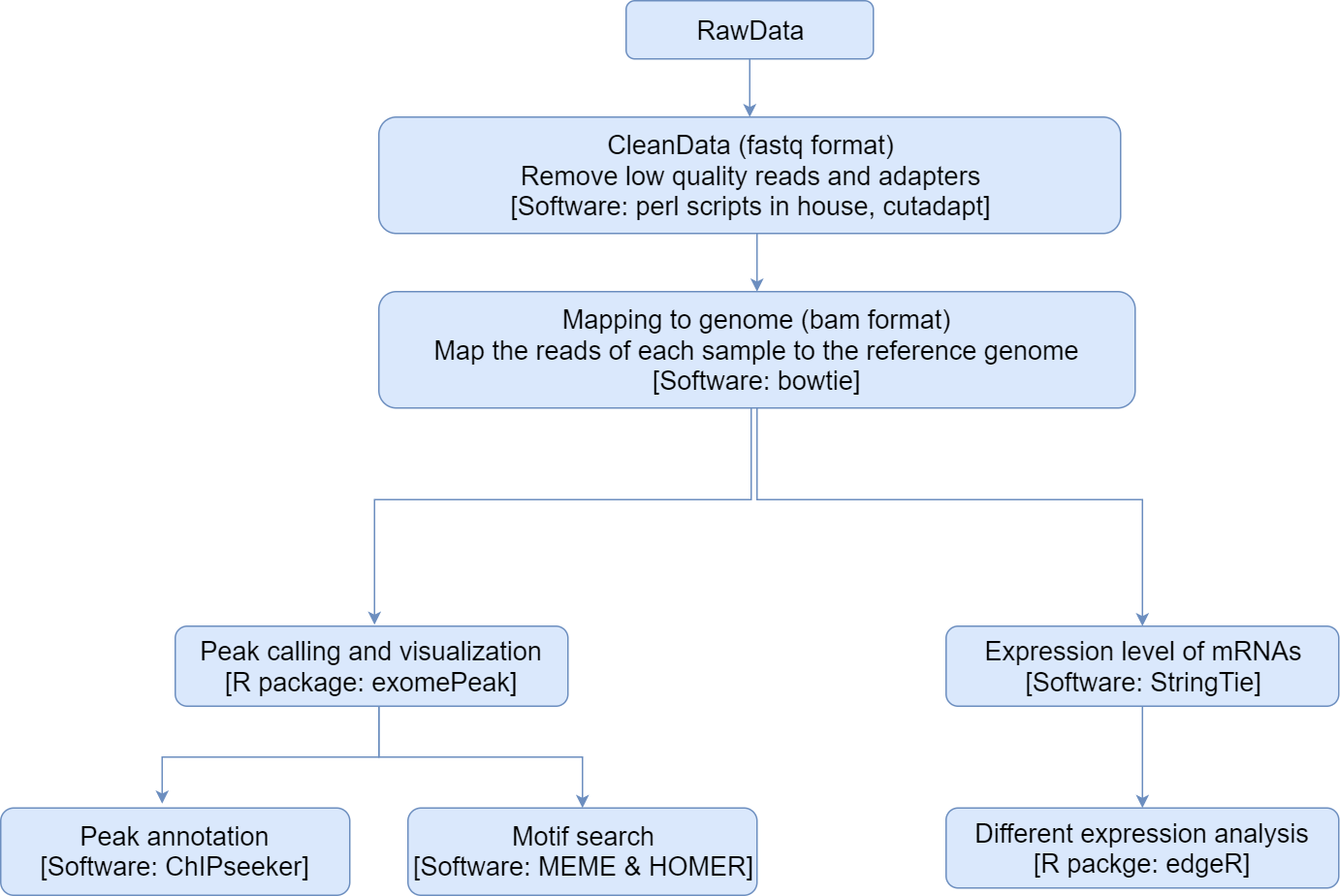
Firstly, Cutadapt[1] and perl scripts in house were used to remove the reads that contained adaptor contamination, low quality bases and undetermined bases. Then sequence quality was verified using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). We used bowtie[2] to map reads to the genome of Homo sapiens (Version: GRCh38.p10) with default parameters. Mapped reads of IP and input libraries were provided for R package exomePeak[3], which identifies m6A peaks with bed or bam format that can be adapted for visualization on the UCSC genome browser or IGV software (http://www.igv.org/). MEME[4] and HOMER[5] were used for de novo and known motif finding followed by localization of the motif with respect to peak summit by perl scripts in house. Called peaks were annotated by intersection with gene architecture using ChIPseeker[6]. Then StringTie[7] was used to perform expression level for all mRNAs from input libraries by calculating FPKM (FPKM=[total_exon_fragments/mapped_reads(millions)×exon_length(kB)]). The differentially expressed mRNAs were selected with log2 (fold change) >1 or log2 (fold change) <-1 and p value < 0.05 by R package edgeR[8].
Reference:
[1] Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet Journal 17 (2011).
[2] Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359 (2012).
[3] Meng, J., et al. A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods. 69(3), 274-281. (2014).
[4] Timothy L. Bailey, et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Research. 37.Web Server issue:202-8. (2009).
[5] Heinz, S., et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and b cell identities. Molecular Cell. 38(4), 576. (2010).
[6] Yu, G., et al. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 31.14:2382. (2015).
[7] Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology. 33, 290–295 (2015).
[8] Robinson, M. D. et al. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26(1), 139. (2010).
Species: Human
Latin name: Homo sapiens
Specimens: METTL14 overexpress HEK293T cell line
| Dababase | Web links | Version/date |
|---|---|---|
| Genome | ftp://ftp.ensembl.org/pub/release-91/fasta/homo_sapiens/dna/ | GRCh38.p10 | Gene Orthology (GO) | http://www.geneontology.org/ | 2016.12 | KEGG | http://www.kegg.jp/ | 2017.06 |
| Analysis item | Software | Version/date | Quality control | FastQC | 0.10.1 | Quality control | Perl scripts in house | NA | Adapter remove | Cutadapt | 1.10 | Genome mapping | bowtie | 1.0 | Trascripts assembly | StringTie | 1.0 | Differential expression analysis | R package: edgeR | 4.1 | Peak Calling | R package: exomePeak | 1.8 | Diff Peak analysis | R package: exomePeak | 1.8 | Peak annotation | CHIPseeker | 1.0 | Motif prediction | MEME | 1.0 | Motif prediction | HOMER | 4.1 | GO and KEGG enrichment analysis | Perl scripts in house | NA |
|---|---|---|
Statistics of reads from 2 libraries was shown below. Results of FastQC, PCA analysis and sample correlation were deposited in summary/3_peak_calling and summary/9_quality_control/.
| Sample_ID | Raw_Reads | Raw_Bases | Valid_Reads | Valid_Bases | Valid% | Q20% | Q30% | GC% |
| M14_NC_IP | 51508042 | 7.78G | 51091328 | 6.74G | 86.67 | 98.87 | 96.62 | 55.99 |
| M14_NC | 67610302 | 10.14G | 65079192 | 9.76G | 96.23 | 97.16 | 92.68 | 52.05 |
| M14_OE_IP | 27012068 | 4.08G | 26816578 | 3.57G | 87.56 | 98.89 | 96.67 | 55.19 |
| M14_OE | 71534530 | 10.73G | 68695446 | 10.30G | 96.00 | 97.05 | 92.42 | 52.20 |
document location: summary/1_raw_data/data_stat.xlsx
| Sample | Valid reads | Mapped reads | Unique Mapped reads | Multi Mapped reads | PE Mapped reads | Reads map to sense strand | Reads map to antisense strand | Non-splice reads |
| M14_NC | 65079192 | 62363860(95.83%) | 27729750(42.61%) | 34634110(53.22%) | 44730600(68.73%) | 14989402(23.03%) | 15020050(23.08%) | 30009452(46.11%) |
| M14_NC_IP | 51091328 | 48456971(94.84%) | 21705470(42.48%) | 26751501(52.36%) | 48241518(94.42%) | 14878094(29.12%) | 14879210(29.12%) | 29757304(58.24%) |
| M14_OE | 68695446 | 66441960(96.72%) | 29479410(42.91%) | 36962550(53.81%) | 48817740(71.06%) | 15966870(23.24%) | 15990275(23.28%) | 31957145(46.52%) |
| M14_OE_IP | 26816578 | 26055929(97.16%) | 11317596(42.20%) | 14738333(54.96%) | 25947782(96.76%) | 7873061(29.36%) | 7873692(29.36%) | 15746753(58.72%) |
document location:summary/2_mapped_stat/mapped_stat_out.xlsx
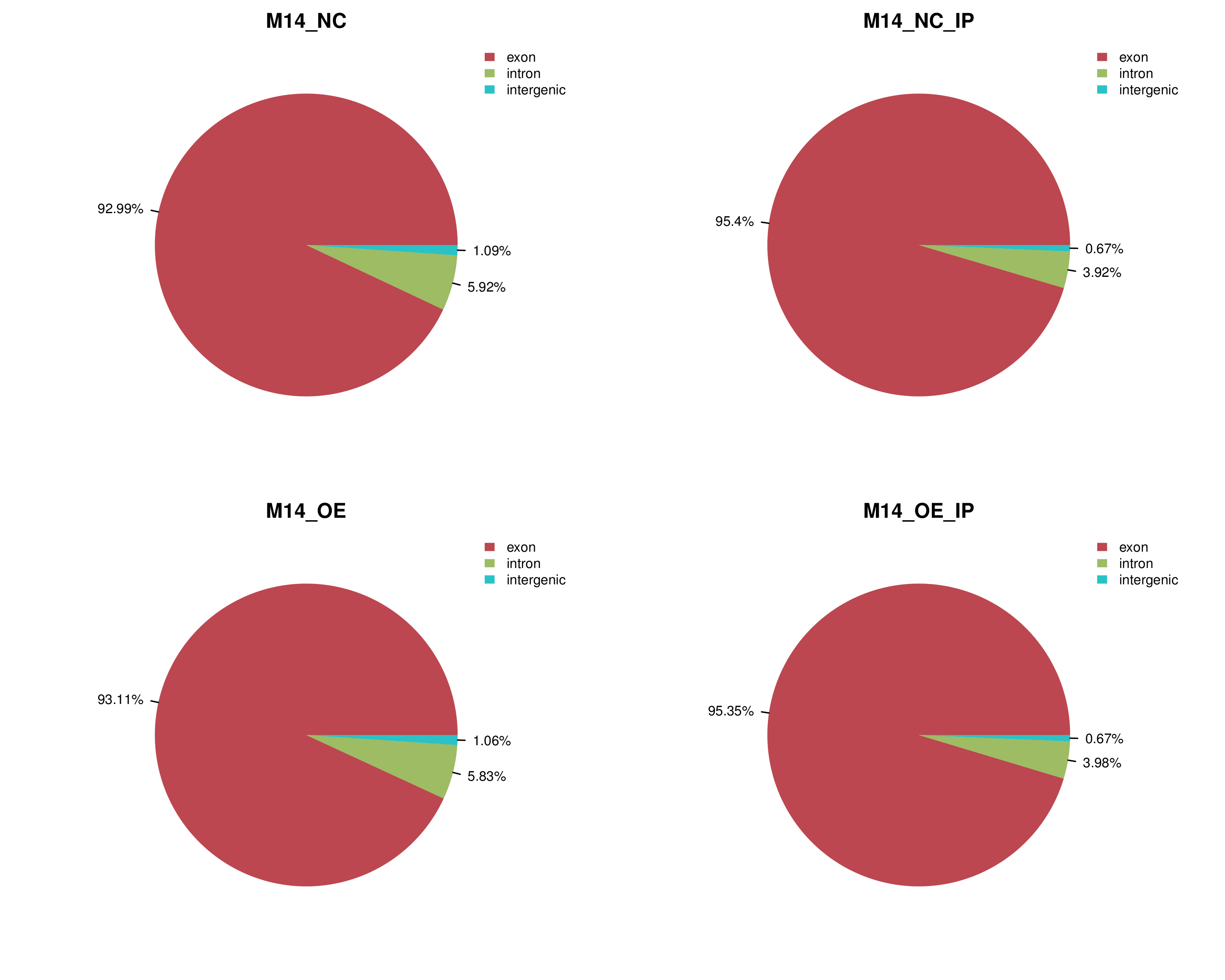
document location:
summary/2_mapped_stat/mapped_region_stat.png
summary/2_mapped_stat/mapped_region_stat.xlsx
The m6A-seq approach is analogous to ChIP-seq—both are based on global identification of regions of signal enrichment. Indeed, the distribution of sequence tags in m6A-seq data is quite similar to that of ChIP-seq: in both cases, enriched regions are discrete and form sharp peaks along the genome or transcriptome. As a marriage of ChIP-Seq and RNA-Seq, m6A-seq has the potential to study the transcriptome-wide distribution of various post-transcriptional RNA modifications. Compared with other relatively well studied data types such as ChIP-Seq and RNA-Seq, the study of m6A-seq data is still at very early stage, and existing protocols are not optimized for dealing with the intrinsic characteristic of m6A-seq data. We utilized an easy-to-use protocol of using exomePeak R/Bioconductor package along with other software programs such as bowtie, ChIPseeker, MEME and HOMER for analysis of m6A-seq data, which covers raw reads alignment, RNA methylation site detection, motif discovery, differential RNA methylation analysis, and functional analysis.
| seqnames | start | end | width | geneId | transcriptId | distanceToTSS | geneName | Description | GO | KEGG | score | blockCount | blockSizes | blockStarts | lg.p | lg.fdr | fold_enrchment | annotation | geneChr | geneStart | geneEnd | geneLength | geneStrand |
| chr1 | 630445 | 630623 | 179 | ENSG00000237973 | ENST00000414273 | -451 | MTCO1P12 | mitochondrially encoded cytochrome c oxidase I pseudogene 12 [Source:HGNC Symbol;Acc:HGNC:52014] | NA | NA | 0.00 | 1 | 179, | 0 | -5.8500000000 | -4.8800000000 | 11.7000000000 | Exon (ENST00000457540/ENSG00000225630, exon 1 of 1) | 1 | 631074 | 632616 | 1543 | 1 |
| chr1 | 924827 | 924887 | 61 | ENSG00000187634 | ENST00000437963 | -263 | SAMD11 | sterile alpha motif domain containing 11 [Source:HGNC Symbol;Acc:HGNC:28706] | GO:0003677(DNA binding);GO:0005634(nucleus) | NA | 0.00 | 1 | 61, | 0 | -2.5600000000 | -1.7500000000 | 4.94 | Exon (ENST00000420190/ENSG00000187634, exon 1 of 7) | 1 | 925150 | 935793 | 10644 | 1 |
| chr1 | 942779 | 942958 | 180 | ENSG00000187634 | ENST00000464948 | 613 | SAMD11 | sterile alpha motif domain containing 11 [Source:HGNC Symbol;Acc:HGNC:28706] | GO:0003677(DNA binding);GO:0005634(nucleus) | NA | 0.00 | 1 | 180, | 0 | -4.1600000000 | -3.2600000000 | 4.55 | 3' UTR | 1 | 942166 | 942892 | 727 | 1 |
| chr1 | 944042 | 944581 | 540 | ENSG00000188976 | ENST00000496938 | 981 | NOC2L | NOC2 like nucleolar associated transcriptional repressor [Source:HGNC Symbol;Acc:HGNC:24517] | GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005730(nucleolus);GO:0005829(cytosol);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0005515(protein binding);GO:0006915(apoptotic process);GO:0005654(nucleoplasm);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0003714(transcription corepressor activity);GO:0003682(chromatin binding);GO:0042273(ribosomal large subunit biogenesis);GO:0042393(histone binding);GO:1901796(regulation of signal transduction by p53 class mediator);GO:0070491(repressing transcription factor binding);GO:2001243(negative regulation of intrinsic apoptotic signaling pathway);GO:0034644(cellular response to UV);GO:0031497(chromatin assembly);GO:0035067(negative regulation of histone acetylation);GO:0002903(negative regulation of B cell apoptotic process);GO:0031491(nucleosome binding);GO:0030690(Noc1p-Noc2p complex);GO:0030691(Noc2p-Noc3p complex) | NA | 0.00 | 1 | 540, | 0 | -52.5000000000 | -50.9000000000 | 8.33 | 3' UTR | 1 | 945319 | 945562 | 244 | 2 |
| chr1 | 965596 | 965715 | 120 | ENSG00000187583 | ENST00000379410 | -782 | PLEKHN1 | pleckstrin homology domain containing N1 [Source:HGNC Symbol;Acc:HGNC:25284] | GO:0016020(membrane);GO:0005886(plasma membrane);GO:0005515(protein binding) | NA | 0.00 | 1 | 120, | 0 | -2.7800000000 | -1.9600000000 | 10.9000000000 | 3' UTR | 1 | 966497 | 975108 | 8612 | 1 |
| chr1 | 974815 | 975865 | 1051 | ENSG00000187642 | ENST00000479361 | 776 | PERM1 | PPARGC1 and ESRR induced regulator, muscle 1 [Source:HGNC Symbol;Acc:HGNC:28208] | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0014850(response to muscle activity) | NA | 0.00 | 2 | 294,154, | 0,897 | -39.1000000000 | -37.6000000000 | 50.8000000000 | 3' UTR | 1 | 975205 | 976641 | 1437 | 2 |
| chr1 | 1014218 | 1014541 | 324 | ENSG00000187608 | ENST00000379389 | 795 | ISG15 | ISG15 ubiquitin-like modifier [Source:HGNC Symbol;Acc:HGNC:4053] | GO:0005737(cytoplasm);GO:0005515(protein binding);GO:0005576(extracellular region);GO:0002376(immune system process);GO:0045087(innate immune response);GO:0042742(defense response to bacterium);GO:0051607(defense response to virus);GO:0005654(nucleoplasm);GO:0005829(cytosol);GO:0045071(negative regulation of viral genome replication);GO:0016032(viral process);GO:0030501(positive regulation of bone mineralization);GO:0031397(negative regulation of protein ubiquitination);GO:0060337(type I interferon signaling pathway);GO:0019985(translesion synthesis);GO:0032480(negative regulation of type I interferon production);GO:0031386(protein tag);GO:0019941(modification-dependent protein catabolic process);GO:0032020(ISG15-protein conjugation);GO:0032649(regulation of interferon-gamma production);GO:0034340(response to type I interferon);GO:0045648(positive regulation of erythrocyte differentiation) | 04622(RIG-I-like receptor signaling pathway);05165(Human papillomavirus infection) | 0.00 | 1 | 324, | 0 | -11.1000000000 | -10 | 4.64 | 3' UTR | 1 | 1013423 | 1014540 | 1118 | 1 |
| chr1 | 1045175 | 1045235 | 61 | ENSG00000188157 | ENST00000479707 | -164 | AGRN | agrin [Source:HGNC Symbol;Acc:HGNC:329] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005829(cytosol);GO:0005509(calcium ion binding);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0007010(cytoskeleton organization);GO:0007165(signal transduction);GO:0005576(extracellular region);GO:0043236(laminin binding);GO:0007275(multicellular organism development);GO:0005578(proteinaceous extracellular matrix);GO:0030054(cell junction);GO:0045202(synapse);GO:0030154(cell differentiation);GO:0005200(structural constituent of cytoskeleton);GO:0007213(G-protein coupled acetylcholine receptor signaling pathway);GO:0043113(receptor clustering);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0070062(extracellular exosome);GO:0043547(positive regulation of GTPase activity);GO:0031012(extracellular matrix);GO:0043202(lysosomal lumen);GO:0002162(dystroglycan binding);GO:0033691(sialic acid binding);GO:0035374(chondroitin sulfate binding);GO:0043395(heparan sulfate proteoglycan binding);GO:0001523(retinoid metabolic process);GO:0006024(glycosaminoglycan biosynthetic process);GO:0006027(glycosaminoglycan catabolic process);GO:0030198(extracellular matrix organization);GO:0030203(glycosaminoglycan metabolic process);GO:0045162(clustering of voltage-gated sodium channels);GO:0045887(positive regulation of synaptic growth at neuromuscular junction);GO:0050808(synapse organization);GO:0051491(positive regulation of filopodium assembly);GO:0005605(basal lamina);GO:0005796(Golgi lumen) | 04512(ECM-receptor interaction) | 0.00 | 1 | 61, | 0 | -6.6300000000 | -5.6300000000 | 4.33 | Exon (ENST00000379370/ENSG00000188157, exon 13 of 36) | 1 | 1045399 | 1046349 | 951 | 1 |
| chr1 | 1046342 | 1046509 | 168 | ENSG00000188157 | ENST00000466223 | -692 | AGRN | agrin [Source:HGNC Symbol;Acc:HGNC:329] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005829(cytosol);GO:0005509(calcium ion binding);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0007010(cytoskeleton organization);GO:0007165(signal transduction);GO:0005576(extracellular region);GO:0043236(laminin binding);GO:0007275(multicellular organism development);GO:0005578(proteinaceous extracellular matrix);GO:0030054(cell junction);GO:0045202(synapse);GO:0030154(cell differentiation);GO:0005200(structural constituent of cytoskeleton);GO:0007213(G-protein coupled acetylcholine receptor signaling pathway);GO:0043113(receptor clustering);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0070062(extracellular exosome);GO:0043547(positive regulation of GTPase activity);GO:0031012(extracellular matrix);GO:0043202(lysosomal lumen);GO:0002162(dystroglycan binding);GO:0033691(sialic acid binding);GO:0035374(chondroitin sulfate binding);GO:0043395(heparan sulfate proteoglycan binding);GO:0001523(retinoid metabolic process);GO:0006024(glycosaminoglycan biosynthetic process);GO:0006027(glycosaminoglycan catabolic process);GO:0030198(extracellular matrix organization);GO:0030203(glycosaminoglycan metabolic process);GO:0045162(clustering of voltage-gated sodium channels);GO:0045887(positive regulation of synaptic growth at neuromuscular junction);GO:0050808(synapse organization);GO:0051491(positive regulation of filopodium assembly);GO:0005605(basal lamina);GO:0005796(Golgi lumen) | 04512(ECM-receptor interaction) | 0.00 | 2 | 8,113, | 0,55 | -13.3000000000 | -12.2000000000 | 3.15 | Exon (ENST00000379370/ENSG00000188157, exon 18 of 36) | 1 | 1047201 | 1047865 | 665 | 1 |
| chr1 | 1048079 | 1048318 | 240 | ENSG00000188157 | ENST00000492947 | -211 | AGRN | agrin [Source:HGNC Symbol;Acc:HGNC:329] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005829(cytosol);GO:0005509(calcium ion binding);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0007010(cytoskeleton organization);GO:0007165(signal transduction);GO:0005576(extracellular region);GO:0043236(laminin binding);GO:0007275(multicellular organism development);GO:0005578(proteinaceous extracellular matrix);GO:0030054(cell junction);GO:0045202(synapse);GO:0030154(cell differentiation);GO:0005200(structural constituent of cytoskeleton);GO:0007213(G-protein coupled acetylcholine receptor signaling pathway);GO:0043113(receptor clustering);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0070062(extracellular exosome);GO:0043547(positive regulation of GTPase activity);GO:0031012(extracellular matrix);GO:0043202(lysosomal lumen);GO:0002162(dystroglycan binding);GO:0033691(sialic acid binding);GO:0035374(chondroitin sulfate binding);GO:0043395(heparan sulfate proteoglycan binding);GO:0001523(retinoid metabolic process);GO:0006024(glycosaminoglycan biosynthetic process);GO:0006027(glycosaminoglycan catabolic process);GO:0030198(extracellular matrix organization);GO:0030203(glycosaminoglycan metabolic process);GO:0045162(clustering of voltage-gated sodium channels);GO:0045887(positive regulation of synaptic growth at neuromuscular junction);GO:0050808(synapse organization);GO:0051491(positive regulation of filopodium assembly);GO:0005605(basal lamina);GO:0005796(Golgi lumen) | 04512(ECM-receptor interaction) | 0.00 | 1 | 240, | 0 | -25.2000000000 | -23.9000000000 | 2.16 | Exon (ENST00000379370/ENSG00000188157, exon 23 of 36) | 1 | 1048529 | 1049394 | 866 | 1 |
| chr1 | 1054979 | 1055249 | 271 | ENSG00000242590 | ENST00000418300 | 0 | AL645608.6 | NA | NA | NA | 0.00 | 1 | 271, | 0 | -105 | -103 | 2.36 | 3' UTR | 1 | 1055033 | 1056116 | 1084 | 1 |
| chr1 | 1055429 | 1056118 | 690 | ENSG00000242590 | ENST00000418300 | 396 | AL645608.6 | NA | NA | NA | 0.00 | 1 | 690, | 0 | -115 | -113 | 10.9000000000 | 3' UTR | 1 | 1055033 | 1056116 | 1084 | 1 |
| chr1 | 1055033 | 1056116 | 1084 | ENSG00000242590 | ENST00000418300 | 0 | AL645608.6 | NA | NA | NA | 0.00 | 2 | 183,219, | 0,865 | -78.4000000000 | -76.7000000000 | 9.99 | 3' UTR | 1 | 1055033 | 1056116 | 1084 | 1 |
| chr1 | 1064506 | 1065447 | 942 | ENSG00000217801 | ENST00000451054 | 0 | AL390719.1 | NA | NA | NA | 0.00 | 2 | 84,96, | 0,846 | -8.1200000000 | -7.0800000000 | 7.65 | Exon (ENST00000412397/ENSG00000217801, exon 4 of 10) | 1 | 1064518 | 1066441 | 1924 | 1 |
| chr1 | 1065860 | 1066276 | 417 | ENSG00000217801 | ENST00000451054 | 1342 | AL390719.1 | NA | NA | NA | 0.00 | 1 | 417, | 0 | -16.9000000000 | -15.7000000000 | 9.37 | Exon (ENST00000427998/ENSG00000217801, exon 4 of 4) | 1 | 1064518 | 1066441 | 1924 | 1 |
| chr1 | 1232623 | 1233519 | 897 | ENSG00000176022 | ENST00000379198 | 358 | B3GALT6 | beta-1,3-galactosyltransferase 6 [Source:HGNC Symbol;Acc:HGNC:17978] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0016740(transferase activity);GO:0016757(transferase activity, transferring glycosyl groups);GO:0006486(protein glycosylation);GO:0005794(Golgi apparatus);GO:0008378(galactosyltransferase activity);GO:0032580(Golgi cisterna membrane);GO:0000139(Golgi membrane);GO:0030203(glycosaminoglycan metabolic process);GO:0015012(heparan sulfate proteoglycan biosynthetic process);GO:0006024(glycosaminoglycan biosynthetic process);GO:0005797(Golgi medial cisterna);GO:0035250(UDP-galactosyltransferase activity);GO:0030206(chondroitin sulfate biosynthetic process);GO:0008499(UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity);GO:0047220(galactosylxylosylprotein 3-beta-galactosyltransferase activity) | 00532(Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate);00534(Glycosaminoglycan biosynthesis - heparan sulfate / heparin) | 0.00 | 1 | 897, | 0 | -20 | -18.7000000000 | 3.92 | 3' UTR | 1 | 1232265 | 1235041 | 2777 | 1 |
| chr1 | 1233668 | 1234235 | 568 | ENSG00000176022 | ENST00000379198 | 1403 | B3GALT6 | beta-1,3-galactosyltransferase 6 [Source:HGNC Symbol;Acc:HGNC:17978] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0016740(transferase activity);GO:0016757(transferase activity, transferring glycosyl groups);GO:0006486(protein glycosylation);GO:0005794(Golgi apparatus);GO:0008378(galactosyltransferase activity);GO:0032580(Golgi cisterna membrane);GO:0000139(Golgi membrane);GO:0030203(glycosaminoglycan metabolic process);GO:0015012(heparan sulfate proteoglycan biosynthetic process);GO:0006024(glycosaminoglycan biosynthetic process);GO:0005797(Golgi medial cisterna);GO:0035250(UDP-galactosyltransferase activity);GO:0030206(chondroitin sulfate biosynthetic process);GO:0008499(UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity);GO:0047220(galactosylxylosylprotein 3-beta-galactosyltransferase activity) | 00532(Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate);00534(Glycosaminoglycan biosynthesis - heparan sulfate / heparin) | 0.00 | 1 | 568, | 0 | -146 | -144 | 7.97 | 3' UTR | 1 | 1232265 | 1235041 | 2777 | 1 |
| chr1 | 1234295 | 1235041 | 747 | ENSG00000176022 | ENST00000379198 | 2030 | B3GALT6 | beta-1,3-galactosyltransferase 6 [Source:HGNC Symbol;Acc:HGNC:17978] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0016740(transferase activity);GO:0016757(transferase activity, transferring glycosyl groups);GO:0006486(protein glycosylation);GO:0005794(Golgi apparatus);GO:0008378(galactosyltransferase activity);GO:0032580(Golgi cisterna membrane);GO:0000139(Golgi membrane);GO:0030203(glycosaminoglycan metabolic process);GO:0015012(heparan sulfate proteoglycan biosynthetic process);GO:0006024(glycosaminoglycan biosynthetic process);GO:0005797(Golgi medial cisterna);GO:0035250(UDP-galactosyltransferase activity);GO:0030206(chondroitin sulfate biosynthetic process);GO:0008499(UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity);GO:0047220(galactosylxylosylprotein 3-beta-galactosyltransferase activity) | 00532(Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate);00534(Glycosaminoglycan biosynthesis - heparan sulfate / heparin) | 0.00 | 1 | 747, | 0 | -59.9000000000 | -58.3000000000 | 17.1000000000 | 3' UTR | 1 | 1232265 | 1235041 | 2777 | 1 |
| chr1 | 1311527 | 1311677 | 151 | ENSG00000283712 | ENST00000620702 | 889 | MIR6727 | microRNA 6727 [Source:HGNC Symbol;Acc:HGNC:50171] | NA | NA | 0.00 | 1 | 151, | 0 | -7.5500000000 | -6.5200000000 | 4.78 | 3' UTR | 1 | 1312502 | 1312566 | 65 | 2 |
| chr1 | 1324994 | 1325280 | 287 | ENSG00000224051 | ENST00000488011 | 0 | CPTP | ceramide-1-phosphate transfer protein [Source:HGNC Symbol;Acc:HGNC:28116] | GO:0006810(transport);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0005829(cytosol);GO:0005794(Golgi apparatus);GO:0005886(plasma membrane);GO:0006869(lipid transport);GO:0120013(intermembrane lipid transfer activity);GO:0120009(intermembrane lipid transfer);GO:0008289(lipid binding);GO:0005768(endosome);GO:0006687(glycosphingolipid metabolic process);GO:0010008(endosome membrane);GO:0005543(phospholipid binding);GO:0005548(phospholipid transporter activity);GO:0005640(nuclear outer membrane);GO:1902387(ceramide 1-phosphate binding);GO:1902388(ceramide 1-phosphate transporter activity);GO:1902389(ceramide 1-phosphate transport) | NA | 0.00 | 2 | 109,40, | 0,247 | -14.2000000000 | -13 | 4.07 | 5' UTR | 1 | 1325241 | 1327471 | 2231 | 1 |
| chr1 | 1327709 | 1328897 | 1189 | ENSG00000169962 | ENST00000339381 | -2417 | TAS1R3 | taste 1 receptor member 3 [Source:HGNC Symbol;Acc:HGNC:15661] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0004930(G-protein coupled receptor activity);GO:0007186(G-protein coupled receptor signaling pathway);GO:0005886(plasma membrane);GO:0004871(signal transducer activity);GO:0007165(signal transduction);GO:0050896(response to stimulus);GO:0050909(sensory perception of taste);GO:0046982(protein heterodimerization activity);GO:0008527(taste receptor activity);GO:0033041(sweet taste receptor activity);GO:0001582(detection of chemical stimulus involved in sensory perception of sweet taste);GO:0050916(sensory perception of sweet taste);GO:0050917(sensory perception of umami taste);GO:1903767(sweet taste receptor complex) | 04973(Carbohydrate digestion and absorption);04742(Taste transduction) | 0.00 | 1 | 1189, | 0 | -198 | -196 | 14.8000000000 | 3' UTR | 1 | 1331314 | 1335306 | 3993 | 1 |
| chr1 | 1332037 | 1332097 | 61 | ENSG00000169962 | ENST00000339381 | 723 | TAS1R3 | taste 1 receptor member 3 [Source:HGNC Symbol;Acc:HGNC:15661] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0004930(G-protein coupled receptor activity);GO:0007186(G-protein coupled receptor signaling pathway);GO:0005886(plasma membrane);GO:0004871(signal transducer activity);GO:0007165(signal transduction);GO:0050896(response to stimulus);GO:0050909(sensory perception of taste);GO:0046982(protein heterodimerization activity);GO:0008527(taste receptor activity);GO:0033041(sweet taste receptor activity);GO:0001582(detection of chemical stimulus involved in sensory perception of sweet taste);GO:0050916(sensory perception of sweet taste);GO:0050917(sensory perception of umami taste);GO:1903767(sweet taste receptor complex) | 04973(Carbohydrate digestion and absorption);04742(Taste transduction) | 0.00 | 1 | 61, | 0 | -2.4300000000 | -1.6300000000 | 10.2000000000 | Exon (ENST00000339381/ENSG00000169962, exon 3 of 6) | 1 | 1331314 | 1335306 | 3993 | 1 |
| chr1 | 1333665 | 1333814 | 150 | ENSG00000169962 | ENST00000339381 | 2351 | TAS1R3 | taste 1 receptor member 3 [Source:HGNC Symbol;Acc:HGNC:15661] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0004930(G-protein coupled receptor activity);GO:0007186(G-protein coupled receptor signaling pathway);GO:0005886(plasma membrane);GO:0004871(signal transducer activity);GO:0007165(signal transduction);GO:0050896(response to stimulus);GO:0050909(sensory perception of taste);GO:0046982(protein heterodimerization activity);GO:0008527(taste receptor activity);GO:0033041(sweet taste receptor activity);GO:0001582(detection of chemical stimulus involved in sensory perception of sweet taste);GO:0050916(sensory perception of sweet taste);GO:0050917(sensory perception of umami taste);GO:1903767(sweet taste receptor complex) | 04973(Carbohydrate digestion and absorption);04742(Taste transduction) | 0.00 | 1 | 150, | 0 | -3.2300000000 | -2.3700000000 | 12.5000000000 | Exon (ENST00000339381/ENSG00000169962, exon 6 of 6) | 1 | 1331314 | 1335306 | 3993 | 1 |
| chr1 | 1334112 | 1334321 | 210 | ENSG00000169962 | ENST00000339381 | 2798 | TAS1R3 | taste 1 receptor member 3 [Source:HGNC Symbol;Acc:HGNC:15661] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0004930(G-protein coupled receptor activity);GO:0007186(G-protein coupled receptor signaling pathway);GO:0005886(plasma membrane);GO:0004871(signal transducer activity);GO:0007165(signal transduction);GO:0050896(response to stimulus);GO:0050909(sensory perception of taste);GO:0046982(protein heterodimerization activity);GO:0008527(taste receptor activity);GO:0033041(sweet taste receptor activity);GO:0001582(detection of chemical stimulus involved in sensory perception of sweet taste);GO:0050916(sensory perception of sweet taste);GO:0050917(sensory perception of umami taste);GO:1903767(sweet taste receptor complex) | 04973(Carbohydrate digestion and absorption);04742(Taste transduction) | 0.00 | 1 | 210, | 0 | -3.2300000000 | -2.3700000000 | 7.42 | Exon (ENST00000339381/ENSG00000169962, exon 6 of 6) | 1 | 1331314 | 1335306 | 3993 | 1 |
| chr1 | 1334381 | 1334441 | 61 | ENSG00000169962 | ENST00000339381 | 3067 | TAS1R3 | taste 1 receptor member 3 [Source:HGNC Symbol;Acc:HGNC:15661] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0004930(G-protein coupled receptor activity);GO:0007186(G-protein coupled receptor signaling pathway);GO:0005886(plasma membrane);GO:0004871(signal transducer activity);GO:0007165(signal transduction);GO:0050896(response to stimulus);GO:0050909(sensory perception of taste);GO:0046982(protein heterodimerization activity);GO:0008527(taste receptor activity);GO:0033041(sweet taste receptor activity);GO:0001582(detection of chemical stimulus involved in sensory perception of sweet taste);GO:0050916(sensory perception of sweet taste);GO:0050917(sensory perception of umami taste);GO:1903767(sweet taste receptor complex) | 04973(Carbohydrate digestion and absorption);04742(Taste transduction) | 0.00 | 1 | 61, | 0 | -2.6800000000 | -1.8600000000 | 10.9000000000 | Exon (ENST00000339381/ENSG00000169962, exon 6 of 6) | 1 | 1331314 | 1335306 | 3993 | 1 |
| chr1 | 1400264 | 1400502 | 239 | ENSG00000224870 | ENST00000453521 | 0 | AL391244.1 | NA | NA | NA | 0.00 | 1 | 239, | 0 | -4.8100000000 | -3.8800000000 | 2.79 | Exon (ENST00000448629/ENSG00000224870, exon 2 of 3) | 1 | 1400419 | 1402046 | 1628 | 1 |
| chr1 | 1400710 | 1402046 | 1337 | ENSG00000224870 | ENST00000453521 | 291 | AL391244.1 | NA | NA | NA | 0.00 | 1 | 1337, | 0 | -42.7000000000 | -41.2000000000 | 19.1000000000 | 3' UTR | 1 | 1400419 | 1402046 | 1628 | 1 |
| chr1 | 1439226 | 1439496 | 271 | ENSG00000179403 | ENST00000471398 | 3491 | VWA1 | von Willebrand factor A domain containing 1 [Source:HGNC Symbol;Acc:HGNC:30910] | GO:0005576(extracellular region);GO:0005578(proteinaceous extracellular matrix);GO:0005615(extracellular space);GO:0005604(basement membrane);GO:0070062(extracellular exosome);GO:0044267(cellular protein metabolic process);GO:0043687(post-translational protein modification);GO:0005788(endoplasmic reticulum lumen);GO:0042802(identical protein binding);GO:0030198(extracellular matrix organization);GO:0048266(behavioral response to pain);GO:0005614(interstitial matrix) | NA | 0.00 | 1 | 271, | 0 | -22.6000000000 | -21.3000000000 | 2.25 | 3' UTR | 1 | 1435735 | 1437177 | 1443 | 1 |
| chr1 | 1439706 | 1440904 | 1199 | ENSG00000179403 | ENST00000471398 | 3971 | VWA1 | von Willebrand factor A domain containing 1 [Source:HGNC Symbol;Acc:HGNC:30910] | GO:0005576(extracellular region);GO:0005578(proteinaceous extracellular matrix);GO:0005615(extracellular space);GO:0005604(basement membrane);GO:0070062(extracellular exosome);GO:0044267(cellular protein metabolic process);GO:0043687(post-translational protein modification);GO:0005788(endoplasmic reticulum lumen);GO:0042802(identical protein binding);GO:0030198(extracellular matrix organization);GO:0048266(behavioral response to pain);GO:0005614(interstitial matrix) | NA | 0 | 1 | 1199, | 0 | -943 | -Inf | 17.1000000000 | 3' UTR | 1 | 1435735 | 1437177 | 1443 | 1 |
| chr1 | 1441743 | 1442313 | 571 | ENSG00000179403 | ENST00000471398 | 6008 | VWA1 | von Willebrand factor A domain containing 1 [Source:HGNC Symbol;Acc:HGNC:30910] | GO:0005576(extracellular region);GO:0005578(proteinaceous extracellular matrix);GO:0005615(extracellular space);GO:0005604(basement membrane);GO:0070062(extracellular exosome);GO:0044267(cellular protein metabolic process);GO:0043687(post-translational protein modification);GO:0005788(endoplasmic reticulum lumen);GO:0042802(identical protein binding);GO:0030198(extracellular matrix organization);GO:0048266(behavioral response to pain);GO:0005614(interstitial matrix) | NA | 0.00 | 1 | 571, | 0 | -23.8000000000 | -22.5000000000 | 5.78 | 3' UTR | 1 | 1435735 | 1437177 | 1443 | 1 |
| chr1 | 1442373 | 1442732 | 360 | ENSG00000179403 | ENST00000471398 | 6638 | VWA1 | von Willebrand factor A domain containing 1 [Source:HGNC Symbol;Acc:HGNC:30910] | GO:0005576(extracellular region);GO:0005578(proteinaceous extracellular matrix);GO:0005615(extracellular space);GO:0005604(basement membrane);GO:0070062(extracellular exosome);GO:0044267(cellular protein metabolic process);GO:0043687(post-translational protein modification);GO:0005788(endoplasmic reticulum lumen);GO:0042802(identical protein binding);GO:0030198(extracellular matrix organization);GO:0048266(behavioral response to pain);GO:0005614(interstitial matrix) | NA | 0.00 | 1 | 360, | 0 | -13.4000000000 | -12.3000000000 | 25 | 3' UTR | 1 | 1435735 | 1437177 | 1443 | 1 |
| chr1 | 1495875 | 1496234 | 360 | ENSG00000284740 | ENST00000641974 | -7016 | AL645728.2 | NA | NA | NA | 0.00 | 1 | 360, | 0 | -30.7000000000 | -29.3000000000 | 46.1000000000 | 3' UTR | 1 | 1503250 | 1509452 | 6203 | 1 |
| chr1 | 1523869 | 1524297 | 429 | ENSG00000197785 | ENST00000400830 | 0 | ATAD3A | ATPase family, AAA domain containing 3A [Source:HGNC Symbol;Acc:HGNC:25567] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0005739(mitochondrion);GO:0005743(mitochondrial inner membrane);GO:0043066(negative regulation of apoptotic process);GO:0016049(cell growth);GO:0042645(mitochondrial nucleoid) | NA | 0.00 | 2 | 96,25, | 0,404 | -4.0200000000 | -3.1200000000 | 15.6000000000 | Exon (ENST00000378756/ENSG00000197785, exon 10 of 16) | 1 | 1524273 | 1529331 | 5059 | 1 |
| chr1 | 1527750 | 1529319 | 1570 | ENSG00000197785 | ENST00000400830 | 3477 | ATAD3A | ATPase family, AAA domain containing 3A [Source:HGNC Symbol;Acc:HGNC:25567] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0005739(mitochondrion);GO:0005743(mitochondrial inner membrane);GO:0043066(negative regulation of apoptotic process);GO:0016049(cell growth);GO:0042645(mitochondrial nucleoid) | NA | 0.00 | 2 | 113,97, | 0,1473 | -3.2600000000 | -2.4000000000 | 4.22 | Exon (ENST00000378756/ENSG00000197785, exon 14 of 16) | 1 | 1524273 | 1529331 | 5059 | 1 |
| chr1 | 1534330 | 1534657 | 328 | ENSG00000205090 | ENST00000624426 | 5693 | TMEM240 | transmembrane protein 240 [Source:HGNC Symbol;Acc:HGNC:25186] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005886(plasma membrane);GO:0030054(cell junction);GO:0045202(synapse);GO:0097060(synaptic membrane) | NA | 0.00 | 1 | 328, | 0 | -17.7000000000 | -16.5000000000 | 5.99 | 3' UTR | 1 | 1539056 | 1540350 | 1295 | 2 |
| chr1 | 1623544 | 1623977 | 434 | ENSG00000197530 | ENST00000508455 | 0 | MIB2 | mindbomb E3 ubiquitin protein ligase 2 [Source:HGNC Symbol;Acc:HGNC:30577] | GO:0005737(cytoplasm);GO:0008270(zinc ion binding);GO:0046872(metal ion binding);GO:0016740(transferase activity);GO:0016567(protein ubiquitination);GO:0003779(actin binding);GO:0004842(ubiquitin-protein transferase activity);GO:0005515(protein binding);GO:0004871(signal transducer activity);GO:0007219(Notch signaling pathway);GO:0005768(endosome);GO:0005829(cytosol);GO:0000209(protein polyubiquitination);GO:0043123(positive regulation of I-kappaB kinase/NF-kappaB signaling);GO:0061630(ubiquitin protein ligase activity);GO:0000151(ubiquitin ligase complex);GO:0005769(early endosome) | NA | 0.00 | 2 | 156,204, | 0,230 | -8.7300000000 | -7.6700000000 | 8 | 3' UTR | 1 | 1623581 | 1624209 | 629 | 1 |
| chr1 | 1624992 | 1625302 | 311 | ENSG00000197530 | ENST00000508148 | 0 | MIB2 | mindbomb E3 ubiquitin protein ligase 2 [Source:HGNC Symbol;Acc:HGNC:30577] | GO:0005737(cytoplasm);GO:0008270(zinc ion binding);GO:0046872(metal ion binding);GO:0016740(transferase activity);GO:0016567(protein ubiquitination);GO:0003779(actin binding);GO:0004842(ubiquitin-protein transferase activity);GO:0005515(protein binding);GO:0004871(signal transducer activity);GO:0007219(Notch signaling pathway);GO:0005768(endosome);GO:0005829(cytosol);GO:0000209(protein polyubiquitination);GO:0043123(positive regulation of I-kappaB kinase/NF-kappaB signaling);GO:0061630(ubiquitin protein ligase activity);GO:0000151(ubiquitin ligase complex);GO:0005769(early endosome) | NA | 0.00 | 2 | 194,17, | 0,294 | -12.4000000000 | -11.2000000000 | 4.64 | 3' UTR | 1 | 1625087 | 1625763 | 677 | 1 |
| chr1 | 1630311 | 1630610 | 300 | ENSG00000197530 | ENST00000470373 | 838 | MIB2 | mindbomb E3 ubiquitin protein ligase 2 [Source:HGNC Symbol;Acc:HGNC:30577] | GO:0005737(cytoplasm);GO:0008270(zinc ion binding);GO:0046872(metal ion binding);GO:0016740(transferase activity);GO:0016567(protein ubiquitination);GO:0003779(actin binding);GO:0004842(ubiquitin-protein transferase activity);GO:0005515(protein binding);GO:0004871(signal transducer activity);GO:0007219(Notch signaling pathway);GO:0005768(endosome);GO:0005829(cytosol);GO:0000209(protein polyubiquitination);GO:0043123(positive regulation of I-kappaB kinase/NF-kappaB signaling);GO:0061630(ubiquitin protein ligase activity);GO:0000151(ubiquitin ligase complex);GO:0005769(early endosome) | NA | 0.00 | 1 | 300, | 0 | -35 | -33.6000000000 | 55.4000000000 | 3' UTR | 1 | 1629473 | 1630605 | 1133 | 1 |
| chr1 | 2150846 | 2156040 | 5195 | ENSG00000067606 | ENST00000478770 | 5459 | PRKCZ | protein kinase C zeta [Source:HGNC Symbol;Acc:HGNC:9412] | GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0000166(nucleotide binding);GO:0004672(protein kinase activity);GO:0004674(protein serine/threonine kinase activity);GO:0005524(ATP binding);GO:0006468(protein phosphorylation);GO:0005829(cytosol);GO:0016740(transferase activity);GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0035556(intracellular signal transduction);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0004697(protein kinase C activity);GO:0030054(cell junction);GO:0006954(inflammatory response);GO:0005768(endosome);GO:0070062(extracellular exosome);GO:0007165(signal transduction);GO:0032753(positive regulation of interleukin-4 production);GO:0051092(positive regulation of NF-kappaB transcription factor activity);GO:0070374(positive regulation of ERK1 and ERK2 cascade);GO:2000667(positive regulation of interleukin-13 secretion);GO:2001181(positive regulation of interleukin-10 secretion);GO:0043066(negative regulation of apoptotic process);GO:0016020(membrane);GO:0031982(vesicle);GO:0005911(cell-cell junction);GO:2000463(positive regulation of excitatory postsynaptic potential);GO:0030010(establishment of cell polarity);GO:0018105(peptidyl-serine phosphorylation);GO:0060291(long-term synaptic potentiation);GO:0007179(transforming growth factor beta receptor signaling pathway);GO:0046627(negative regulation of insulin receptor signaling pathway);GO:0043560(insulin receptor substrate binding);GO:0050732(negative regulation of peptidyl-tyrosine phosphorylation);GO:2000664(positive regulation of interleukin-5 secretion);GO:0045630(positive regulation of T-helper 2 cell differentiation);GO:0031333(negative regulation of protein complex assembly);GO:0015459(potassium channel regulator activity);GO:0019901(protein kinase binding);GO:0019904(protein domain specific binding);GO:0043274(phospholipase binding);GO:0071889(14-3-3 protein binding);GO:0000226(microtubule cytoskeleton organization);GO:0001954(positive regulation of cell-matrix adhesion);GO:0007166(cell surface receptor signaling pathway);GO:0007616(long-term memory);GO:0008284(positive regulation of cell proliferation);GO:0008286(insulin receptor signaling pathway);GO:0016477(cell migration);GO:0031532(actin cytoskeleton reorganization);GO:0031584(activation of phospholipase D activity);GO:0032148(activation of protein kinase B activity);GO:0032869(cellular response to insulin stimulus);GO:0034613(cellular protein localization);GO:0046326(positive regulation of glucose import);GO:0046628(positive regulation of insulin receptor signaling pathway);GO:0047496(vesicle transport along microtubule);GO:0050806(positive regulation of synaptic transmission);GO:0051222(positive regulation of protein transport);GO:0051291(protein heterooligomerization);GO:0051346(negative regulation of hydrolase activity);GO:0051899(membrane depolarization);GO:0060081(membrane hyperpolarization);GO:0070528(protein kinase C signaling);GO:0072659(protein localization to plasma membrane);GO:1990138(neuron projection extension);GO:2000553(positive regulation of T-helper 2 cell cytokine production);GO:0005634(nucleus);GO:0005635(nuclear envelope);GO:0005815(microtubule organizing center);GO:0005923(bicellular tight junction);GO:0005938(cell cortex);GO:0016324(apical plasma membrane);GO:0016363(nuclear matrix);GO:0031252(cell leading edge);GO:0031941(filamentous actin);GO:0035748(myelin sheath abaxonal region);GO:0043203(axon hillock);GO:0043231(intracellular membrane-bounded organelle);GO:0043234(protein complex);GO:0045121(membrane raft);GO:0045179(apical cortex);GO:0048471(perinuclear region of cytoplasm);GO:0005622(intracellular) | 04015(Rap1 signaling pathway);04390(Hippo signaling pathway);04071(Sphingolipid signaling pathway);04144(Endocytosis);04530(Tight junction);04611(Platelet activation);04062(Chemokine signaling pathway);04910(Insulin signaling pathway);04926(Relaxin signaling pathway);04360(Axon guidance);05418(Fluid shear stress and atherosclerosis);04930(Type II diabetes mellitus);04931(Insulin resistance);04933(AGE-RAGE signaling pathway in diabetic complications);05165(Human papillomavirus infection) | 0.00 | 2 | 134,46, | 0,5149 | -3.8400000000 | -2.9500000000 | 2.56 | 3' UTR | 1 | 2145387 | 2185116 | 39730 | 1 |
| chr1 | 2184603 | 2185395 | 793 | ENSG00000182873 | ENST00000444529 | -214 | PRKCZ-AS1 | PRKCZ antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:40477] | NA | NA | 0.00 | 2 | 96,474, | 0,319 | -36.7000000000 | -35.3000000000 | 6.16 | 3' UTR | 1 | 2182735 | 2184389 | 1655 | 2 |
| chr1 | 2229474 | 2232516 | 3043 | ENSG00000157933 | ENST00000508416 | 0 | SKI | SKI proto-oncogene [Source:HGNC Symbol;Acc:HGNC:10896] | GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0005737(cytoplasm);GO:0048147(negative regulation of fibroblast proliferation);GO:0007179(transforming growth factor beta receptor signaling pathway);GO:0005634(nucleus);GO:0005654(nucleoplasm);GO:0008270(zinc ion binding);GO:0006351(transcription, DNA-templated);GO:0005515(protein binding);GO:0016604(nuclear body);GO:0046332(SMAD binding);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0003714(transcription corepressor activity);GO:0019901(protein kinase binding);GO:0008285(negative regulation of cell proliferation);GO:0031625(ubiquitin protein ligase binding);GO:0043234(protein complex);GO:0005667(transcription factor complex);GO:0019904(protein domain specific binding);GO:0030509(BMP signaling pathway);GO:0070491(repressing transcription factor binding);GO:0016605(PML body);GO:0060395(SMAD protein signal transduction);GO:0060021(palate development);GO:0030177(positive regulation of Wnt signaling pathway);GO:0001843(neural tube closure);GO:0005813(centrosome);GO:0022011(myelination in peripheral nervous system);GO:0030512(negative regulation of transforming growth factor beta receptor signaling pathway);GO:0017053(transcriptional repressor complex);GO:0030514(negative regulation of BMP signaling pathway);GO:0045668(negative regulation of osteoblast differentiation);GO:0043388(positive regulation of DNA binding);GO:0060349(bone morphogenesis);GO:0032926(negative regulation of activin receptor signaling pathway);GO:0009948(anterior/posterior axis specification);GO:0008283(cell proliferation);GO:0043585(nose morphogenesis);GO:0060325(face morphogenesis);GO:0070207(protein homotrimerization);GO:0030326(embryonic limb morphogenesis);GO:0046811(histone deacetylase inhibitor activity);GO:0002089(lens morphogenesis in camera-type eye);GO:0010626(negative regulation of Schwann cell proliferation);GO:0014902(myotube differentiation);GO:0021772(olfactory bulb development);GO:0031064(negative regulation of histone deacetylation);GO:0035019(somatic stem cell population maintenance);GO:0043010(camera-type eye development);GO:0048593(camera-type eye morphogenesis);GO:0048666(neuron development);GO:0048741(skeletal muscle fiber development);GO:0048870(cell motility);GO:0060041(retina development in camera-type eye) | NA | 0.00 | 2 | 262,38, | 0,3005 | -17.8000000000 | -16.6000000000 | 3.83 | Exon (ENST00000378536/ENSG00000157933, exon 1 of 7) | 1 | 2232479 | 2303038 | 70560 | 1 |
| chr1 | 2306050 | 2307454 | 1405 | ENSG00000157933 | ENST00000507179 | 2404 | SKI | SKI proto-oncogene [Source:HGNC Symbol;Acc:HGNC:10896] | GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0005737(cytoplasm);GO:0048147(negative regulation of fibroblast proliferation);GO:0007179(transforming growth factor beta receptor signaling pathway);GO:0005634(nucleus);GO:0005654(nucleoplasm);GO:0008270(zinc ion binding);GO:0006351(transcription, DNA-templated);GO:0005515(protein binding);GO:0016604(nuclear body);GO:0046332(SMAD binding);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0003714(transcription corepressor activity);GO:0019901(protein kinase binding);GO:0008285(negative regulation of cell proliferation);GO:0031625(ubiquitin protein ligase binding);GO:0043234(protein complex);GO:0005667(transcription factor complex);GO:0019904(protein domain specific binding);GO:0030509(BMP signaling pathway);GO:0070491(repressing transcription factor binding);GO:0016605(PML body);GO:0060395(SMAD protein signal transduction);GO:0060021(palate development);GO:0030177(positive regulation of Wnt signaling pathway);GO:0001843(neural tube closure);GO:0005813(centrosome);GO:0022011(myelination in peripheral nervous system);GO:0030512(negative regulation of transforming growth factor beta receptor signaling pathway);GO:0017053(transcriptional repressor complex);GO:0030514(negative regulation of BMP signaling pathway);GO:0045668(negative regulation of osteoblast differentiation);GO:0043388(positive regulation of DNA binding);GO:0060349(bone morphogenesis);GO:0032926(negative regulation of activin receptor signaling pathway);GO:0009948(anterior/posterior axis specification);GO:0008283(cell proliferation);GO:0043585(nose morphogenesis);GO:0060325(face morphogenesis);GO:0070207(protein homotrimerization);GO:0030326(embryonic limb morphogenesis);GO:0046811(histone deacetylase inhibitor activity);GO:0002089(lens morphogenesis in camera-type eye);GO:0010626(negative regulation of Schwann cell proliferation);GO:0014902(myotube differentiation);GO:0021772(olfactory bulb development);GO:0031064(negative regulation of histone deacetylation);GO:0035019(somatic stem cell population maintenance);GO:0043010(camera-type eye development);GO:0048593(camera-type eye morphogenesis);GO:0048666(neuron development);GO:0048741(skeletal muscle fiber development);GO:0048870(cell motility);GO:0060041(retina development in camera-type eye) | NA | 0.00 | 2 | 201,878, | 0,527 | -128 | -126 | 10.8000000000 | 3' UTR | 1 | 2303646 | 2304368 | 723 | 1 |
| chr1 | 2307663 | 2308023 | 361 | ENSG00000157933 | ENST00000507179 | 4017 | SKI | SKI proto-oncogene [Source:HGNC Symbol;Acc:HGNC:10896] | GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0005737(cytoplasm);GO:0048147(negative regulation of fibroblast proliferation);GO:0007179(transforming growth factor beta receptor signaling pathway);GO:0005634(nucleus);GO:0005654(nucleoplasm);GO:0008270(zinc ion binding);GO:0006351(transcription, DNA-templated);GO:0005515(protein binding);GO:0016604(nuclear body);GO:0046332(SMAD binding);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0003714(transcription corepressor activity);GO:0019901(protein kinase binding);GO:0008285(negative regulation of cell proliferation);GO:0031625(ubiquitin protein ligase binding);GO:0043234(protein complex);GO:0005667(transcription factor complex);GO:0019904(protein domain specific binding);GO:0030509(BMP signaling pathway);GO:0070491(repressing transcription factor binding);GO:0016605(PML body);GO:0060395(SMAD protein signal transduction);GO:0060021(palate development);GO:0030177(positive regulation of Wnt signaling pathway);GO:0001843(neural tube closure);GO:0005813(centrosome);GO:0022011(myelination in peripheral nervous system);GO:0030512(negative regulation of transforming growth factor beta receptor signaling pathway);GO:0017053(transcriptional repressor complex);GO:0030514(negative regulation of BMP signaling pathway);GO:0045668(negative regulation of osteoblast differentiation);GO:0043388(positive regulation of DNA binding);GO:0060349(bone morphogenesis);GO:0032926(negative regulation of activin receptor signaling pathway);GO:0009948(anterior/posterior axis specification);GO:0008283(cell proliferation);GO:0043585(nose morphogenesis);GO:0060325(face morphogenesis);GO:0070207(protein homotrimerization);GO:0030326(embryonic limb morphogenesis);GO:0046811(histone deacetylase inhibitor activity);GO:0002089(lens morphogenesis in camera-type eye);GO:0010626(negative regulation of Schwann cell proliferation);GO:0014902(myotube differentiation);GO:0021772(olfactory bulb development);GO:0031064(negative regulation of histone deacetylation);GO:0035019(somatic stem cell population maintenance);GO:0043010(camera-type eye development);GO:0048593(camera-type eye morphogenesis);GO:0048666(neuron development);GO:0048741(skeletal muscle fiber development);GO:0048870(cell motility);GO:0060041(retina development in camera-type eye) | NA | 0.00 | 1 | 361, | 0 | -21.6000000000 | -20.3000000000 | 2.88 | 3' UTR | 1 | 2303646 | 2304368 | 723 | 1 |
| chr1 | 2309849 | 2310119 | 271 | ENSG00000157933 | ENST00000507179 | 6203 | SKI | SKI proto-oncogene [Source:HGNC Symbol;Acc:HGNC:10896] | GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0005737(cytoplasm);GO:0048147(negative regulation of fibroblast proliferation);GO:0007179(transforming growth factor beta receptor signaling pathway);GO:0005634(nucleus);GO:0005654(nucleoplasm);GO:0008270(zinc ion binding);GO:0006351(transcription, DNA-templated);GO:0005515(protein binding);GO:0016604(nuclear body);GO:0046332(SMAD binding);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0003714(transcription corepressor activity);GO:0019901(protein kinase binding);GO:0008285(negative regulation of cell proliferation);GO:0031625(ubiquitin protein ligase binding);GO:0043234(protein complex);GO:0005667(transcription factor complex);GO:0019904(protein domain specific binding);GO:0030509(BMP signaling pathway);GO:0070491(repressing transcription factor binding);GO:0016605(PML body);GO:0060395(SMAD protein signal transduction);GO:0060021(palate development);GO:0030177(positive regulation of Wnt signaling pathway);GO:0001843(neural tube closure);GO:0005813(centrosome);GO:0022011(myelination in peripheral nervous system);GO:0030512(negative regulation of transforming growth factor beta receptor signaling pathway);GO:0017053(transcriptional repressor complex);GO:0030514(negative regulation of BMP signaling pathway);GO:0045668(negative regulation of osteoblast differentiation);GO:0043388(positive regulation of DNA binding);GO:0060349(bone morphogenesis);GO:0032926(negative regulation of activin receptor signaling pathway);GO:0009948(anterior/posterior axis specification);GO:0008283(cell proliferation);GO:0043585(nose morphogenesis);GO:0060325(face morphogenesis);GO:0070207(protein homotrimerization);GO:0030326(embryonic limb morphogenesis);GO:0046811(histone deacetylase inhibitor activity);GO:0002089(lens morphogenesis in camera-type eye);GO:0010626(negative regulation of Schwann cell proliferation);GO:0014902(myotube differentiation);GO:0021772(olfactory bulb development);GO:0031064(negative regulation of histone deacetylation);GO:0035019(somatic stem cell population maintenance);GO:0043010(camera-type eye development);GO:0048593(camera-type eye morphogenesis);GO:0048666(neuron development);GO:0048741(skeletal muscle fiber development);GO:0048870(cell motility);GO:0060041(retina development in camera-type eye) | NA | 0.00 | 1 | 271, | 0 | -20.8000000000 | -19.5000000000 | 47.6000000000 | 3' UTR | 1 | 2303646 | 2304368 | 723 | 1 |
| chr1 | 2391895 | 2393230 | 1336 | ENSG00000157916 | ENST00000443438 | 0 | RER1 | retention in endoplasmic reticulum sorting receptor 1 [Source:HGNC Symbol;Acc:HGNC:30309] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005794(Golgi apparatus);GO:0000139(Golgi membrane);GO:0005886(plasma membrane);GO:0003674(molecular_function);GO:0006890(retrograde vesicle-mediated transport, Golgi to ER);GO:0030173(integral component of Golgi membrane);GO:0033130(acetylcholine receptor binding);GO:0007528(neuromuscular junction development);GO:0071340(skeletal muscle acetylcholine-gated channel clustering);GO:0090004(positive regulation of establishment of protein localization to plasma membrane);GO:0005793(endoplasmic reticulum-Golgi intermediate compartment);GO:0009986(cell surface) | NA | 0.00 | 2 | 64,57, | 0,1279 | -4.5200000000 | -3.6000000000 | 2.71 | 5' UTR | 1 | 2393174 | 2402316 | 9143 | 1 |
| chr1 | 2403048 | 2403168 | 121 | ENSG00000157916 | ENST00000462129 | 1097 | RER1 | retention in endoplasmic reticulum sorting receptor 1 [Source:HGNC Symbol;Acc:HGNC:30309] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005794(Golgi apparatus);GO:0000139(Golgi membrane);GO:0005886(plasma membrane);GO:0003674(molecular_function);GO:0006890(retrograde vesicle-mediated transport, Golgi to ER);GO:0030173(integral component of Golgi membrane);GO:0033130(acetylcholine receptor binding);GO:0007528(neuromuscular junction development);GO:0071340(skeletal muscle acetylcholine-gated channel clustering);GO:0090004(positive regulation of establishment of protein localization to plasma membrane);GO:0005793(endoplasmic reticulum-Golgi intermediate compartment);GO:0009986(cell surface) | NA | 0.00 | 1 | 121, | 0 | -6.5100000000 | -5.5100000000 | 1.49 | 3' UTR | 1 | 2401951 | 2403227 | 1277 | 1 |
| chr1 | 2403348 | 2403767 | 420 | ENSG00000157916 | ENST00000462129 | 1397 | RER1 | retention in endoplasmic reticulum sorting receptor 1 [Source:HGNC Symbol;Acc:HGNC:30309] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005794(Golgi apparatus);GO:0000139(Golgi membrane);GO:0005886(plasma membrane);GO:0003674(molecular_function);GO:0006890(retrograde vesicle-mediated transport, Golgi to ER);GO:0030173(integral component of Golgi membrane);GO:0033130(acetylcholine receptor binding);GO:0007528(neuromuscular junction development);GO:0071340(skeletal muscle acetylcholine-gated channel clustering);GO:0090004(positive regulation of establishment of protein localization to plasma membrane);GO:0005793(endoplasmic reticulum-Golgi intermediate compartment);GO:0009986(cell surface) | NA | 0.00 | 1 | 420, | 0 | -49.2000000000 | -47.6000000000 | 4.02 | 3' UTR | 1 | 2401951 | 2403227 | 1277 | 1 |
| chr1 | 2404875 | 2405444 | 570 | ENSG00000157916 | ENST00000462129 | 2924 | RER1 | retention in endoplasmic reticulum sorting receptor 1 [Source:HGNC Symbol;Acc:HGNC:30309] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005794(Golgi apparatus);GO:0000139(Golgi membrane);GO:0005886(plasma membrane);GO:0003674(molecular_function);GO:0006890(retrograde vesicle-mediated transport, Golgi to ER);GO:0030173(integral component of Golgi membrane);GO:0033130(acetylcholine receptor binding);GO:0007528(neuromuscular junction development);GO:0071340(skeletal muscle acetylcholine-gated channel clustering);GO:0090004(positive regulation of establishment of protein localization to plasma membrane);GO:0005793(endoplasmic reticulum-Golgi intermediate compartment);GO:0009986(cell surface) | NA | 0.00 | 1 | 570, | 0 | -39 | -37.5000000000 | 2.54 | 3' UTR | 1 | 2401951 | 2403227 | 1277 | 1 |
| chr1 | 2504093 | 2504183 | 91 | ENSG00000149527 | ENST00000462379 | 5446 | PLCH2 | phospholipase C eta 2 [Source:HGNC Symbol;Acc:HGNC:29037] | GO:0005622(intracellular);GO:0035556(intracellular signal transduction);GO:0004629(phospholipase C activity);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0046872(metal ion binding);GO:0016787(hydrolase activity);GO:0005509(calcium ion binding);GO:0007165(signal transduction);GO:0006629(lipid metabolic process);GO:0016042(lipid catabolic process);GO:0005886(plasma membrane);GO:0004871(signal transducer activity);GO:0008081(phosphoric diester hydrolase activity);GO:0004435(phosphatidylinositol phospholipase C activity);GO:0003674(molecular_function);GO:0008150(biological_process);GO:0005575(cellular_component);GO:0043647(inositol phosphate metabolic process);GO:0046488(phosphatidylinositol metabolic process) | 00562(Inositol phosphate metabolism) | 0.00 | 1 | 91, | 0 | -2.8800000000 | -2.0400000000 | 11.7000000000 | 3' UTR | 1 | 2498647 | 2505528 | 6882 | 1 |
| chr1 | 2546845 | 2547401 | 557 | ENSG00000272449 | ENST00000606645 | 380 | AL139246.5 | NA | NA | NA | 0.00 | 1 | 557, | 0 | -9.7100000000 | -8.6300000000 | 34.4000000000 | Exon (ENST00000606645/ENSG00000272449, exon 1 of 1) | 1 | 2546465 | 2547460 | 996 | 1 |
| chr1 | 2563113 | 2563352 | 240 | ENSG00000157873 | ENST00000480305 | 1312 | TNFRSF14 | TNF receptor superfamily member 14 [Source:HGNC Symbol;Acc:HGNC:11912] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0002741(positive regulation of cytokine secretion involved in immune response);GO:0046642(negative regulation of alpha-beta T cell proliferation);GO:0050731(positive regulation of peptidyl-tyrosine phosphorylation);GO:0050829(defense response to Gram-negative bacterium);GO:0050830(defense response to Gram-positive bacterium);GO:2000406(positive regulation of T cell migration);GO:0009897(external side of plasma membrane);GO:0005515(protein binding);GO:0005886(plasma membrane);GO:0005887(integral component of plasma membrane);GO:0006954(inflammatory response);GO:0046718(viral entry into host cell);GO:0097190(apoptotic signaling pathway);GO:0033209(tumor necrosis factor-mediated signaling pathway);GO:0016032(viral process);GO:0031625(ubiquitin protein ligase binding);GO:0006955(immune response);GO:0007275(multicellular organism development);GO:0032496(response to lipopolysaccharide);GO:0042127(regulation of cell proliferation);GO:0007166(cell surface receptor signaling pathway);GO:0042981(regulation of apoptotic process);GO:0001618(virus receptor activity);GO:0031295(T cell costimulation);GO:0005031(tumor necrosis factor-activated receptor activity) | 04060(Cytokine-cytokine receptor interaction);05168(Herpes simplex infection) | 0.00 | 1 | 240, | 0 | -14.9000000000 | -13.7000000000 | 3.76 | 3' UTR | 1 | 2561801 | 2563452 | 1652 | 1 |
| chr1 | 2563591 | 2565083 | 1493 | ENSG00000225931 | ENST00000456687 | -1327 | AL139246.2 | NA | NA | NA | 0.00 | 2 | 239,31, | 0,1462 | -31.9000000000 | -30.5000000000 | 25.5000000000 | 3' UTR | 1 | 2566410 | 2569888 | 3479 | 1 |
| chr1 | 2586798 | 2587100 | 303 | ENSG00000157870 | ENST00000378427 | 0 | FAM213B | family with sequence similarity 213 member B [Source:HGNC Symbol;Acc:HGNC:28390] | GO:0005737(cytoplasm);GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0006629(lipid metabolic process);GO:0005829(cytosol);GO:0005783(endoplasmic reticulum);GO:0006631(fatty acid metabolic process);GO:0006633(fatty acid biosynthetic process);GO:0098869(cellular oxidant detoxification);GO:0070062(extracellular exosome);GO:0001516(prostaglandin biosynthetic process);GO:0006693(prostaglandin metabolic process);GO:0043209(myelin sheath);GO:0047017(prostaglandin-F synthase activity);GO:0016209(antioxidant activity);GO:0016616(oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor) | 00590(Arachidonic acid metabolism) | 0.00 | 2 | 151,59, | 0,244 | -24.9000000000 | -23.6000000000 | 8.15 | Exon (ENST00000493183/ENSG00000157870, exon 2 of 7) | 1 | 2586798 | 2591463 | 4666 | 1 |
| chr1 | 2589411 | 2589680 | 270 | ENSG00000157870 | ENST00000476686 | 817 | FAM213B | family with sequence similarity 213 member B [Source:HGNC Symbol;Acc:HGNC:28390] | GO:0005737(cytoplasm);GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0006629(lipid metabolic process);GO:0005829(cytosol);GO:0005783(endoplasmic reticulum);GO:0006631(fatty acid metabolic process);GO:0006633(fatty acid biosynthetic process);GO:0098869(cellular oxidant detoxification);GO:0070062(extracellular exosome);GO:0001516(prostaglandin biosynthetic process);GO:0006693(prostaglandin metabolic process);GO:0043209(myelin sheath);GO:0047017(prostaglandin-F synthase activity);GO:0016209(antioxidant activity);GO:0016616(oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor) | 00590(Arachidonic acid metabolism) | 0.00 | 1 | 270, | 0 | -25.6000000000 | -24.2000000000 | 2.33 | 3' UTR | 1 | 2588594 | 2591468 | 2875 | 1 |
| chr1 | 2590455 | 2590544 | 90 | ENSG00000157870 | ENST00000476686 | 1861 | FAM213B | family with sequence similarity 213 member B [Source:HGNC Symbol;Acc:HGNC:28390] | GO:0005737(cytoplasm);GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0006629(lipid metabolic process);GO:0005829(cytosol);GO:0005783(endoplasmic reticulum);GO:0006631(fatty acid metabolic process);GO:0006633(fatty acid biosynthetic process);GO:0098869(cellular oxidant detoxification);GO:0070062(extracellular exosome);GO:0001516(prostaglandin biosynthetic process);GO:0006693(prostaglandin metabolic process);GO:0043209(myelin sheath);GO:0047017(prostaglandin-F synthase activity);GO:0016209(antioxidant activity);GO:0016616(oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor) | 00590(Arachidonic acid metabolism) | 0.00 | 1 | 90, | 0 | -4.4700000000 | -3.5500000000 | 1.45 | 3' UTR | 1 | 2588594 | 2591468 | 2875 | 1 |
| chr1 | 2591022 | 2591469 | 448 | ENSG00000142606 | ENST00000471840 | 1209 | MMEL1 | membrane metalloendopeptidase like 1 [Source:HGNC Symbol;Acc:HGNC:14668] | GO:0006508(proteolysis);GO:0004222(metalloendopeptidase activity);GO:0008237(metallopeptidase activity);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0046872(metal ion binding);GO:0016787(hydrolase activity);GO:0008233(peptidase activity);GO:0005576(extracellular region) | NA | 0.00 | 1 | 448, | 0 | -182 | -180 | 97.6000000000 | 3' UTR | 1 | 2590639 | 2592678 | 2040 | 2 |
| chr1 | 3454695 | 3454785 | 91 | ENSG00000130762 | ENST00000378378 | 269 | ARHGEF16 | Rho guanine nucleotide exchange factor 16 [Source:HGNC Symbol;Acc:HGNC:15515] | GO:0005737(cytoplasm);GO:0005089(Rho guanyl-nucleotide exchange factor activity);GO:0035023(regulation of Rho protein signal transduction);GO:0005515(protein binding);GO:0005085(guanyl-nucleotide exchange factor activity);GO:0005829(cytosol);GO:0045296(cadherin binding);GO:0051056(regulation of small GTPase mediated signal transduction);GO:0090630(activation of GTPase activity);GO:0030971(receptor tyrosine kinase binding);GO:0060326(cell chemotaxis);GO:0043065(positive regulation of apoptotic process);GO:0017048(Rho GTPase binding);GO:0030165(PDZ domain binding);GO:0090004(positive regulation of establishment of protein localization to plasma membrane);GO:0043547(positive regulation of GTPase activity) | NA | 0.00 | 1 | 91, | 0 | -2.9500000000 | -2.1100000000 | 5.47 | 5' UTR | 1 | 3454426 | 3481113 | 26688 | 1 |
| chr1 | 3463248 | 3463577 | 330 | ENSG00000130762 | ENST00000378373 | -2428 | ARHGEF16 | Rho guanine nucleotide exchange factor 16 [Source:HGNC Symbol;Acc:HGNC:15515] | GO:0005737(cytoplasm);GO:0005089(Rho guanyl-nucleotide exchange factor activity);GO:0035023(regulation of Rho protein signal transduction);GO:0005515(protein binding);GO:0005085(guanyl-nucleotide exchange factor activity);GO:0005829(cytosol);GO:0045296(cadherin binding);GO:0051056(regulation of small GTPase mediated signal transduction);GO:0090630(activation of GTPase activity);GO:0030971(receptor tyrosine kinase binding);GO:0060326(cell chemotaxis);GO:0043065(positive regulation of apoptotic process);GO:0017048(Rho GTPase binding);GO:0030165(PDZ domain binding);GO:0090004(positive regulation of establishment of protein localization to plasma membrane);GO:0043547(positive regulation of GTPase activity) | NA | 0.00 | 1 | 330, | 0 | -6.1400000000 | -5.1500000000 | 3.42 | Exon (ENST00000378378/ENSG00000130762, exon 2 of 15) | 1 | 3466005 | 3481113 | 15109 | 1 |
| chr1 | 3480784 | 3481053 | 270 | ENSG00000272088 | ENST00000606489 | -6193 | AL512413.1 | NA | NA | NA | 0.00 | 1 | 270, | 0 | -3.4300000000 | -2.5700000000 | 11.7000000000 | 3' UTR | 1 | 3487246 | 3487627 | 382 | 1 |
| chr1 | 3625002 | 3625450 | 449 | ENSG00000158109 | ENST00000378344 | 0 | TPRG1L | tumor protein p63 regulated 1 like [Source:HGNC Symbol;Acc:HGNC:27007] | GO:0016020(membrane);GO:0030054(cell junction);GO:0045202(synapse);GO:0031410(cytoplasmic vesicle);GO:0003674(molecular_function);GO:0008150(biological_process);GO:0070062(extracellular exosome);GO:0030672(synaptic vesicle membrane);GO:0008021(synaptic vesicle);GO:0042802(identical protein binding) | NA | 0.00 | 2 | 272,27, | 0,422 | -15.2000000000 | -14 | 36.7000000000 | 5' UTR | 1 | 3625002 | 3630127 | 5126 | 1 |
| chr1 | 3628786 | 3628935 | 150 | ENSG00000158109 | ENST00000344579 | 3736 | TPRG1L | tumor protein p63 regulated 1 like [Source:HGNC Symbol;Acc:HGNC:27007] | GO:0016020(membrane);GO:0030054(cell junction);GO:0045202(synapse);GO:0031410(cytoplasmic vesicle);GO:0003674(molecular_function);GO:0008150(biological_process);GO:0070062(extracellular exosome);GO:0030672(synaptic vesicle membrane);GO:0008021(synaptic vesicle);GO:0042802(identical protein binding) | NA | 0.00 | 1 | 150, | 0 | -5.4000000000 | -4.4400000000 | 1.77 | 3' UTR | 1 | 3625050 | 3630127 | 5078 | 1 |
| chr1 | 3629948 | 3630127 | 180 | ENSG00000158109 | ENST00000344579 | 4898 | TPRG1L | tumor protein p63 regulated 1 like [Source:HGNC Symbol;Acc:HGNC:27007] | GO:0016020(membrane);GO:0030054(cell junction);GO:0045202(synapse);GO:0031410(cytoplasmic vesicle);GO:0003674(molecular_function);GO:0008150(biological_process);GO:0070062(extracellular exosome);GO:0030672(synaptic vesicle membrane);GO:0008021(synaptic vesicle);GO:0042802(identical protein binding) | NA | 0.00 | 1 | 180, | 0 | -9.9600000000 | -8.8700000000 | 16.4000000000 | 3' UTR | 1 | 3625050 | 3630127 | 5078 | 1 |
| chr1 | 3707607 | 3707757 | 151 | ENSG00000235131 | ENST00000416554 | 6541 | AL136528.2 | NA | NA | NA | 0.00 | 1 | 151, | 0 | -3.6100000000 | -2.7400000000 | 5.27 | Exon (ENST00000378295/ENSG00000078900, exon 4 of 14) | 1 | 3712200 | 3714298 | 2099 | 2 |
| chr1 | 3733508 | 3733748 | 241 | ENSG00000227372 | ENST00000624167 | 2840 | TP73-AS1 | TP73 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:29052] | NA | NA | 0.00 | 1 | 241, | 0 | -7.1000000000 | -6.0900000000 | 5.27 | 3' UTR | 1 | 3735601 | 3736588 | 988 | 2 |
| chr1 | 3746610 | 3751344 | 4735 | ENSG00000227372 | ENST00000418088 | 0 | TP73-AS1 | TP73 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:29052] | NA | NA | 0.00 | 3 | 82,145,13, | 0,1204,4722 | -17.4000000000 | -16.2000000000 | 15.2000000000 | Exon (ENST00000636250/ENSG00000162592, exon 1 of 6) | 1 | 3735984 | 3746776 | 10793 | 2 |
| chr1 | 3785096 | 3785244 | 149 | ENSG00000272153 | ENST00000607459 | 88 | AL365330.1 | NA | NA | NA | 0.00 | 1 | 149, | 0 | -4.2100000000 | -3.3000000000 | 5.86 | Exon (ENST00000607459/ENSG00000272153, exon 1 of 1) | 1 | 3785008 | 3785538 | 531 | 1 |
| chr1 | 6098495 | 6098795 | 301 | ENSG00000069424 | ENST00000481789 | 2239 | KCNAB2 | potassium voltage-gated channel subfamily A regulatory beta subunit 2 [Source:HGNC Symbol;Acc:HGNC:6229] | GO:0016021(integral component of membrane);GO:0005249(voltage-gated potassium channel activity);GO:0006813(potassium ion transport);GO:0071805(potassium ion transmembrane transport);GO:0006810(transport);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0005829(cytosol);GO:0005244(voltage-gated ion channel activity);GO:0006811(ion transport);GO:0034765(regulation of ion transmembrane transport);GO:0005886(plasma membrane);GO:0005856(cytoskeleton);GO:0042995(cell projection);GO:0065009(regulation of molecular function);GO:0030054(cell junction);GO:0045202(synapse);GO:0005874(microtubule);GO:0030424(axon);GO:0031234(extrinsic component of cytoplasmic side of plasma membrane);GO:0043312(neutrophil degranulation);GO:0035579(specific granule membrane);GO:0070821(tertiary granule membrane);GO:0015459(potassium channel regulator activity);GO:0043005(neuron projection);GO:2000008(regulation of protein localization to cell surface);GO:0008076(voltage-gated potassium channel complex);GO:0004033(aldo-keto reductase (NADP) activity);GO:0044224(juxtaparanode region of axon);GO:1901379(regulation of potassium ion transmembrane transport);GO:0070995(NADPH oxidation);GO:1990031(pinceau fiber);GO:0002244(hematopoietic progenitor cell differentiation);GO:0050905(neuromuscular process);GO:0014069(postsynaptic density);GO:0043679(axon terminus) | NA | 0.00 | 1 | 301, | 0 | -41.7000000000 | -40.2000000000 | 3.71 | 3' UTR | 1 | 6096256 | 6098810 | 2555 | 1 |
| chr1 | 6099214 | 6099304 | 91 | ENSG00000069424 | ENST00000481789 | 2958 | KCNAB2 | potassium voltage-gated channel subfamily A regulatory beta subunit 2 [Source:HGNC Symbol;Acc:HGNC:6229] | GO:0016021(integral component of membrane);GO:0005249(voltage-gated potassium channel activity);GO:0006813(potassium ion transport);GO:0071805(potassium ion transmembrane transport);GO:0006810(transport);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0005829(cytosol);GO:0005244(voltage-gated ion channel activity);GO:0006811(ion transport);GO:0034765(regulation of ion transmembrane transport);GO:0005886(plasma membrane);GO:0005856(cytoskeleton);GO:0042995(cell projection);GO:0065009(regulation of molecular function);GO:0030054(cell junction);GO:0045202(synapse);GO:0005874(microtubule);GO:0030424(axon);GO:0031234(extrinsic component of cytoplasmic side of plasma membrane);GO:0043312(neutrophil degranulation);GO:0035579(specific granule membrane);GO:0070821(tertiary granule membrane);GO:0015459(potassium channel regulator activity);GO:0043005(neuron projection);GO:2000008(regulation of protein localization to cell surface);GO:0008076(voltage-gated potassium channel complex);GO:0004033(aldo-keto reductase (NADP) activity);GO:0044224(juxtaparanode region of axon);GO:1901379(regulation of potassium ion transmembrane transport);GO:0070995(NADPH oxidation);GO:1990031(pinceau fiber);GO:0002244(hematopoietic progenitor cell differentiation);GO:0050905(neuromuscular process);GO:0014069(postsynaptic density);GO:0043679(axon terminus) | NA | 0.00 | 1 | 91, | 0 | -2.8300000000 | -2 | 2.09 | 3' UTR | 1 | 6096256 | 6098810 | 2555 | 1 |
| chr1 | 6099544 | 6099694 | 151 | ENSG00000069424 | ENST00000481789 | 3288 | KCNAB2 | potassium voltage-gated channel subfamily A regulatory beta subunit 2 [Source:HGNC Symbol;Acc:HGNC:6229] | GO:0016021(integral component of membrane);GO:0005249(voltage-gated potassium channel activity);GO:0006813(potassium ion transport);GO:0071805(potassium ion transmembrane transport);GO:0006810(transport);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0005829(cytosol);GO:0005244(voltage-gated ion channel activity);GO:0006811(ion transport);GO:0034765(regulation of ion transmembrane transport);GO:0005886(plasma membrane);GO:0005856(cytoskeleton);GO:0042995(cell projection);GO:0065009(regulation of molecular function);GO:0030054(cell junction);GO:0045202(synapse);GO:0005874(microtubule);GO:0030424(axon);GO:0031234(extrinsic component of cytoplasmic side of plasma membrane);GO:0043312(neutrophil degranulation);GO:0035579(specific granule membrane);GO:0070821(tertiary granule membrane);GO:0015459(potassium channel regulator activity);GO:0043005(neuron projection);GO:2000008(regulation of protein localization to cell surface);GO:0008076(voltage-gated potassium channel complex);GO:0004033(aldo-keto reductase (NADP) activity);GO:0044224(juxtaparanode region of axon);GO:1901379(regulation of potassium ion transmembrane transport);GO:0070995(NADPH oxidation);GO:1990031(pinceau fiber);GO:0002244(hematopoietic progenitor cell differentiation);GO:0050905(neuromuscular process);GO:0014069(postsynaptic density);GO:0043679(axon terminus) | NA | 0.00 | 1 | 151, | 0 | -7.0900000000 | -6.0800000000 | 2.94 | 3' UTR | 1 | 6096256 | 6098810 | 2555 | 1 |
| chr1 | 6100923 | 6101193 | 271 | ENSG00000069424 | ENST00000481789 | 4667 | KCNAB2 | potassium voltage-gated channel subfamily A regulatory beta subunit 2 [Source:HGNC Symbol;Acc:HGNC:6229] | GO:0016021(integral component of membrane);GO:0005249(voltage-gated potassium channel activity);GO:0006813(potassium ion transport);GO:0071805(potassium ion transmembrane transport);GO:0006810(transport);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0005829(cytosol);GO:0005244(voltage-gated ion channel activity);GO:0006811(ion transport);GO:0034765(regulation of ion transmembrane transport);GO:0005886(plasma membrane);GO:0005856(cytoskeleton);GO:0042995(cell projection);GO:0065009(regulation of molecular function);GO:0030054(cell junction);GO:0045202(synapse);GO:0005874(microtubule);GO:0030424(axon);GO:0031234(extrinsic component of cytoplasmic side of plasma membrane);GO:0043312(neutrophil degranulation);GO:0035579(specific granule membrane);GO:0070821(tertiary granule membrane);GO:0015459(potassium channel regulator activity);GO:0043005(neuron projection);GO:2000008(regulation of protein localization to cell surface);GO:0008076(voltage-gated potassium channel complex);GO:0004033(aldo-keto reductase (NADP) activity);GO:0044224(juxtaparanode region of axon);GO:1901379(regulation of potassium ion transmembrane transport);GO:0070995(NADPH oxidation);GO:1990031(pinceau fiber);GO:0002244(hematopoietic progenitor cell differentiation);GO:0050905(neuromuscular process);GO:0014069(postsynaptic density);GO:0043679(axon terminus) | NA | 0.00 | 1 | 271, | 0 | -33.7000000000 | -32.3000000000 | 22.9000000000 | 3' UTR | 1 | 6096256 | 6098810 | 2555 | 1 |
| chr1 | 6580882 | 6581091 | 210 | ENSG00000204859 | ENST00000488936 | 794 | ZBTB48 | zinc finger and BTB domain containing 48 [Source:HGNC Symbol;Acc:HGNC:4930] | GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0005694(chromosome);GO:0005515(protein binding);GO:0045893(positive regulation of transcription, DNA-templated);GO:0042802(identical protein binding);GO:0000781(chromosome, telomeric region);GO:0044212(transcription regulatory region DNA binding);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0003691(double-stranded telomeric DNA binding);GO:0010833(telomere maintenance via telomere lengthening) | NA | 0.00 | 1 | 210, | 0 | -14.6000000000 | -13.5000000000 | 5.14 | Exon (ENST00000319084/ENSG00000204859, exon 2 of 3) | 1 | 6580088 | 6588105 | 8018 | 1 |
| chr1 | 6588982 | 6589280 | 299 | ENSG00000204859 | ENST00000466941 | 295 | ZBTB48 | zinc finger and BTB domain containing 48 [Source:HGNC Symbol;Acc:HGNC:4930] | GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0005694(chromosome);GO:0005515(protein binding);GO:0045893(positive regulation of transcription, DNA-templated);GO:0042802(identical protein binding);GO:0000781(chromosome, telomeric region);GO:0044212(transcription regulatory region DNA binding);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0003691(double-stranded telomeric DNA binding);GO:0010833(telomere maintenance via telomere lengthening) | NA | 0.00 | 1 | 299, | 0 | -12 | -10.9000000000 | 28.9000000000 | 3' UTR | 1 | 6588687 | 6589280 | 594 | 1 |
| chr1 | 6613835 | 6614104 | 270 | ENSG00000116273 | ENST00000495385 | 73 | PHF13 | PHD finger protein 13 [Source:HGNC Symbol;Acc:HGNC:22983] | GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0016569(covalent chromatin modification);GO:0051301(cell division);GO:0007049(cell cycle);GO:0005654(nucleoplasm);GO:0007059(chromosome segregation);GO:0003682(chromatin binding);GO:0030261(chromosome condensation);GO:0000278(mitotic cell cycle);GO:0035064(methylated histone binding);GO:0007076(mitotic chromosome condensation) | NA | 0.00 | 1 | 270, | 0 | -18.2000000000 | -17 | 6.07 | 5' UTR | 1 | 6613762 | 6622645 | 8884 | 1 |
| chr1 | 6619938 | 6620297 | 360 | ENSG00000041988 | ENST00000054650 | -4569 | THAP3 | THAP domain containing 3 [Source:HGNC Symbol;Acc:HGNC:20855] | GO:0003676(nucleic acid binding);GO:0046872(metal ion binding);GO:0003677(DNA binding);GO:0005515(protein binding) | NA | 0.00 | 1 | 360, | 0 | -11.1000000000 | -9.9900000000 | 2.37 | Exon (ENST00000377648/ENSG00000116273, exon 3 of 4) | 1 | 6624866 | 6633565 | 8700 | 1 |
| chr1 | 6621490 | 6621909 | 420 | ENSG00000041988 | ENST00000054650 | -2957 | THAP3 | THAP domain containing 3 [Source:HGNC Symbol;Acc:HGNC:20855] | GO:0003676(nucleic acid binding);GO:0046872(metal ion binding);GO:0003677(DNA binding);GO:0005515(protein binding) | NA | 0.00 | 1 | 420, | 0 | -32.9000000000 | -31.5000000000 | 4.78 | 3' UTR | 1 | 6624866 | 6633565 | 8700 | 1 |
| chr1 | 6632887 | 6633516 | 630 | ENSG00000041988 | ENST00000480647 | 4492 | THAP3 | THAP domain containing 3 [Source:HGNC Symbol;Acc:HGNC:20855] | GO:0003676(nucleic acid binding);GO:0046872(metal ion binding);GO:0003677(DNA binding);GO:0005515(protein binding) | NA | 0.00 | 1 | 630, | 0 | -66.6000000000 | -64.9000000000 | 16.4000000000 | 3' UTR | 1 | 6628395 | 6630326 | 1932 | 1 |
| chr1 | 6634239 | 6634628 | 390 | ENSG00000041988 | ENST00000480647 | 5844 | THAP3 | THAP domain containing 3 [Source:HGNC Symbol;Acc:HGNC:20855] | GO:0003676(nucleic acid binding);GO:0046872(metal ion binding);GO:0003677(DNA binding);GO:0005515(protein binding) | NA | 0.00 | 1 | 390, | 0 | -20.1000000000 | -18.8000000000 | 5.40 | 3' UTR | 1 | 6628395 | 6630326 | 1932 | 1 |
| chr1 | 6785504 | 6807001 | 21498 | ENSG00000171735 | ENST00000467404 | 0 | CAMTA1 | calmodulin binding transcription activator 1 [Source:HGNC Symbol;Acc:HGNC:18806] | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0005730(nucleolus);GO:0005829(cytosol);GO:0006366(transcription from RNA polymerase II promoter);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0001077(transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding);GO:0043565(sequence-specific DNA binding);GO:0050885(neuromuscular process controlling balance) | NA | 0.00 | 2 | 72,19, | 0,21479 | -3.1000000000 | -2.2500000000 | 12.5000000000 | 5' UTR | 1 | 6785518 | 6872019 | 86502 | 1 |
| chr1 | 7732491 | 7732580 | 90 | ENSG00000171735 | ENST00000495233 | -3828 | CAMTA1 | calmodulin binding transcription activator 1 [Source:HGNC Symbol;Acc:HGNC:18806] | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0005730(nucleolus);GO:0005829(cytosol);GO:0006366(transcription from RNA polymerase II promoter);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0001077(transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding);GO:0043565(sequence-specific DNA binding);GO:0050885(neuromuscular process controlling balance) | NA | 0.00 | 1 | 90, | 0 | -2.6500000000 | -1.8300000000 | 10.9000000000 | Exon (ENST00000303635/ENSG00000171735, exon 12 of 23) | 1 | 7736408 | 7767856 | 31449 | 1 |
| chr1 | 7772740 | 7772920 | 181 | ENSG00000049245 | ENST00000470357 | 33 | VAMP3 | vesicle associated membrane protein 3 [Source:HGNC Symbol;Acc:HGNC:12644] | GO:0006810(transport);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0015031(protein transport);GO:0016192(vesicle-mediated transport);GO:0005829(cytosol);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0030054(cell junction);GO:0045202(synapse);GO:0006906(vesicle fusion);GO:0043231(intracellular membrane-bounded organelle);GO:0005622(intracellular);GO:0061024(membrane organization);GO:0002479(antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent);GO:0042147(retrograde transport, endosome to Golgi);GO:0000149(SNARE binding);GO:0005484(SNAP receptor activity);GO:0061025(membrane fusion);GO:0030670(phagocytic vesicle membrane);GO:0031201(SNARE complex);GO:0043005(neuron projection);GO:0032588(trans-Golgi network membrane);GO:0030136(clathrin-coated vesicle);GO:0006461(protein complex assembly);GO:0030133(transport vesicle);GO:0006887(exocytosis);GO:0030665(clathrin-coated vesicle membrane);GO:0001921(positive regulation of receptor recycling);GO:0055038(recycling endosome membrane);GO:0034446(substrate adhesion-dependent cell spreading);GO:0055037(recycling endosome);GO:0017075(syntaxin-1 binding);GO:0006904(vesicle docking involved in exocytosis);GO:0017156(calcium ion regulated exocytosis);GO:0035493(SNARE complex assembly);GO:0043001(Golgi to plasma membrane protein transport);GO:1903531(negative regulation of secretion by cell);GO:0009986(cell surface);GO:0016324(apical plasma membrane);GO:0030141(secretory granule);GO:0031410(cytoplasmic vesicle);GO:0043229(intracellular organelle);GO:0048471(perinuclear region of cytoplasm) | 04130(SNARE interactions in vesicular transport);04145(Phagosome) | 0.00 | 1 | 181, | 0 | -7.6300000000 | -6.6000000000 | 4.51 | 5' UTR | 1 | 7772707 | 7779806 | 7100 | 1 |
| chr1 | 7962840 | 7965378 | 2539 | ENSG00000116288 | ENST00000469225 | 0 | PARK7 | Parkinsonism associated deglycase [Source:HGNC Symbol;Acc:HGNC:16369] | GO:0048471(perinuclear region of cytoplasm);GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0005829(cytosol);GO:0016787(hydrolase activity);GO:0005739(mitochondrion);GO:0042803(protein homodimerization activity);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0008233(peptidase activity);GO:0006508(proteolysis);GO:0006914(autophagy);GO:0098793(presynapse);GO:0045121(membrane raft);GO:0098869(cellular oxidant detoxification);GO:0006954(inflammatory response);GO:0042802(identical protein binding);GO:0003729(mRNA binding);GO:0045296(cadherin binding);GO:0070062(extracellular exosome);GO:0050821(protein stabilization);GO:0007338(single fertilization);GO:0005102(receptor binding);GO:0005913(cell-cell adherens junction);GO:0005507(copper ion binding);GO:0051897(positive regulation of protein kinase B signaling);GO:0032091(negative regulation of protein binding);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0008134(transcription factor binding);GO:0007005(mitochondrion organization);GO:1903206(negative regulation of hydrogen peroxide-induced cell death);GO:2001237(negative regulation of extrinsic apoptotic signaling pathway);GO:0019900(kinase binding);GO:0051091(positive regulation of sequence-specific DNA binding transcription factor activity);GO:0034599(cellular response to oxidative stress);GO:1901215(negative regulation of neuron death);GO:0000785(chromatin);GO:0060548(negative regulation of cell death);GO:1902236(negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway);GO:0019899(enzyme binding);GO:1990381(ubiquitin-specific protease binding);GO:0010628(positive regulation of gene expression);GO:0010629(negative regulation of gene expression);GO:0043524(negative regulation of neuron apoptotic process);GO:1903202(negative regulation of oxidative stress-induced cell death);GO:1903377(negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway);GO:1903599(positive regulation of mitophagy);GO:0006517(protein deglycosylation);GO:0050727(regulation of inflammatory response);GO:0070301(cellular response to hydrogen peroxide);GO:0051444(negative regulation of ubiquitin-protein transferase activity);GO:0031397(negative regulation of protein ubiquitination);GO:0043066(negative regulation of apoptotic process);GO:0032148(activation of protein kinase B activity);GO:0033138(positive regulation of peptidyl-serine phosphorylation);GO:0030424(axon);GO:0060765(regulation of androgen receptor signaling pathway);GO:2000825(positive regulation of androgen receptor activity);GO:1905259(negative regulation of nitrosative stress-induced intrinsic apoptotic signaling pathway);GO:0051881(regulation of mitochondrial membrane potential);GO:0005747(mitochondrial respiratory chain complex I);GO:1903208(negative regulation of hydrogen peroxide-induced neuron death);GO:1903094(negative regulation of protein K48-linked deubiquitination);GO:2000157(negative regulation of ubiquitin-specific protease activity);GO:0010273(detoxification of copper ion);GO:0070491(repressing transcription factor binding);GO:0043523(regulation of neuron apoptotic process);GO:0097110(scaffold protein binding);GO:0001933(negative regulation of protein phosphorylation);GO:0044390(ubiquitin-like protein conjugating enzyme binding);GO:0003713(transcription coactivator activity);GO:0016532(superoxide dismutase copper chaperone activity);GO:0016684(oxidoreductase activity, acting on peroxide as acceptor);GO:0019955(cytokine binding);GO:0036470(tyrosine 3-monooxygenase activator activity);GO:0036478(L-dopa decarboxylase activator activity);GO:0036524(protein deglycase activity);GO:0044388(small protein activating enzyme binding);GO:0045340(mercury ion binding);GO:0050681(androgen receptor binding);GO:1903135(cupric ion binding);GO:1903136(cuprous ion binding);GO:0002866(positive regulation of acute inflammatory response to antigenic stimulus);GO:0006469(negative regulation of protein kinase activity);GO:0007265(Ras protein signal transduction);GO:0009438(methylglyoxal metabolic process);GO:0019249(lactate biosynthetic process);GO:0032435(negative regulation of proteasomal ubiquitin-dependent protein catabolic process);GO:0032757(positive regulation of interleukin-8 production);GO:0033234(negative regulation of protein sumoylation);GO:0033864(positive regulation of NAD(P)H oxidase activity);GO:0036471(cellular response to glyoxal);GO:0036526(peptidyl-cysteine deglycation);GO:0036527(peptidyl-arginine deglycation);GO:0036528(peptidyl-lysine deglycation);GO:0036529(protein deglycation, glyoxal removal);GO:0036530(protein deglycation, methylglyoxal removal);GO:0036531(glutathione deglycation);GO:0042743(hydrogen peroxide metabolic process);GO:0046295(glycolate biosynthetic process);GO:0046826(negative regulation of protein export from nucleus);GO:0050787(detoxification of mercury ion);GO:0090073(positive regulation of protein homodimerization activity);GO:1900182(positive regulation of protein localization to nucleus);GO:1901671(positive regulation of superoxide dismutase activity);GO:1901984(negative regulation of protein acetylation);GO:1902903(regulation of supramolecular fiber organization);GO:1902958(positive regulation of mitochondrial electron transport, NADH to ubiquinone);GO:1903073(negative regulation of death-inducing signaling complex assembly);GO:1903122(negative regulation of TRAIL-activated apoptotic signaling pathway);GO:1903168(positive regulation of pyrroline-5-carboxylate reductase activity);GO:1903178(positive regulation of tyrosine 3-monooxygenase activity);GO:1903181(positive regulation of dopamine biosynthetic process);GO:1903189(glyoxal metabolic process);GO:1903197(positive regulation of L-dopa biosynthetic process);GO:1903200(positive regulation of L-dopa decarboxylase activity);GO:1903384(negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway);GO:2000679(positive regulation of transcription regulatory region DNA binding);GO:2001268(negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway);GO:0016605(PML body);GO:0051920(peroxiredoxin activity);GO:0001963(synaptic transmission, dopaminergic);GO:0008344(adult locomotory behavior);GO:0018323(enzyme active site formation via L-cysteine sulfinic acid);GO:0034614(cellular response to reactive oxygen species);GO:0042177(negative regulation of protein catabolic process);GO:0042542(response to hydrogen peroxide);GO:0051583(dopamine uptake involved in synaptic transmission);GO:0051899(membrane depolarization);GO:0060081(membrane hyperpolarization);GO:1902177(positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway);GO:1903204(negative regulation of oxidative stress-induced neuron death);GO:1903428(positive regulation of reactive oxygen species biosynthetic process);GO:2000277(positive regulation of oxidative phosphorylation uncoupler activity);GO:0005758(mitochondrial intermembrane space);GO:0005759(mitochondrial matrix);GO:0005783(endoplasmic reticulum);GO:0043005(neuron projection);GO:0044297(cell body) | 05012(Parkinson's disease) | 0.00 | 2 | 36,55, | 0,2484 | -3.5600000000 | -2.6800000000 | 14.1000000000 | Exon (ENST00000493373/ENSG00000116288, exon 2 of 7) | 1 | 7965351 | 7985282 | 19932 | 1 |
| chr1 | 7984876 | 7985146 | 271 | ENSG00000200344 | ENST00000363474 | -1893 | Y_RNA | Y RNA [Source:RFAM;Acc:RF00019] | NA | NA | 0.00 | 1 | 271, | 0 | -33.2000000000 | -31.8000000000 | 3.24 | 3' UTR | 1 | 7982881 | 7982983 | 103 | 2 |
| chr1 | 9245237 | 9245387 | 151 | ENSG00000049239 | ENST00000602477 | 5393 | H6PD | hexose-6-phosphate dehydrogenase/glucose 1-dehydrogenase [Source:HGNC Symbol;Acc:HGNC:4795] | GO:0016491(oxidoreductase activity);GO:0050661(NADP binding);GO:0055114(oxidation-reduction process);GO:0016787(hydrolase activity);GO:0005975(carbohydrate metabolic process);GO:0005783(endoplasmic reticulum);GO:0003824(catalytic activity);GO:0008152(metabolic process);GO:0005788(endoplasmic reticulum lumen);GO:0006006(glucose metabolic process);GO:0017057(6-phosphogluconolactonase activity);GO:0006098(pentose-phosphate shunt);GO:0004345(glucose-6-phosphate dehydrogenase activity);GO:0047936(glucose 1-dehydrogenase [NAD(P)] activity);GO:0030246(carbohydrate binding);GO:0006739(NADP metabolic process);GO:0097305(response to alcohol);GO:0005737(cytoplasm);GO:0005739(mitochondrion);GO:0043231(intracellular membrane-bounded organelle) | 00030(Pentose phosphate pathway) | 0.00 | 1 | 151, | 0 | -3.4100000000 | -2.5500000000 | 2.11 | Exon (ENST00000377403/ENSG00000049239, exon 2 of 5) | 1 | 9239844 | 9267898 | 28055 | 1 |
| chr1 | 9264594 | 9264654 | 61 | ENSG00000049239 | ENST00000495451 | -876 | H6PD | hexose-6-phosphate dehydrogenase/glucose 1-dehydrogenase [Source:HGNC Symbol;Acc:HGNC:4795] | GO:0016491(oxidoreductase activity);GO:0050661(NADP binding);GO:0055114(oxidation-reduction process);GO:0016787(hydrolase activity);GO:0005975(carbohydrate metabolic process);GO:0005783(endoplasmic reticulum);GO:0003824(catalytic activity);GO:0008152(metabolic process);GO:0005788(endoplasmic reticulum lumen);GO:0006006(glucose metabolic process);GO:0017057(6-phosphogluconolactonase activity);GO:0006098(pentose-phosphate shunt);GO:0004345(glucose-6-phosphate dehydrogenase activity);GO:0047936(glucose 1-dehydrogenase [NAD(P)] activity);GO:0030246(carbohydrate binding);GO:0006739(NADP metabolic process);GO:0097305(response to alcohol);GO:0005737(cytoplasm);GO:0005739(mitochondrion);GO:0043231(intracellular membrane-bounded organelle) | 00030(Pentose phosphate pathway) | 0.00 | 1 | 61, | 0 | -2.5700000000 | -1.7500000000 | 2.04 | Exon (ENST00000377403/ENSG00000049239, exon 5 of 5) | 1 | 9265530 | 9271337 | 5808 | 1 |
| chr1 | 9264864 | 9264924 | 61 | ENSG00000049239 | ENST00000495451 | -606 | H6PD | hexose-6-phosphate dehydrogenase/glucose 1-dehydrogenase [Source:HGNC Symbol;Acc:HGNC:4795] | GO:0016491(oxidoreductase activity);GO:0050661(NADP binding);GO:0055114(oxidation-reduction process);GO:0016787(hydrolase activity);GO:0005975(carbohydrate metabolic process);GO:0005783(endoplasmic reticulum);GO:0003824(catalytic activity);GO:0008152(metabolic process);GO:0005788(endoplasmic reticulum lumen);GO:0006006(glucose metabolic process);GO:0017057(6-phosphogluconolactonase activity);GO:0006098(pentose-phosphate shunt);GO:0004345(glucose-6-phosphate dehydrogenase activity);GO:0047936(glucose 1-dehydrogenase [NAD(P)] activity);GO:0030246(carbohydrate binding);GO:0006739(NADP metabolic process);GO:0097305(response to alcohol);GO:0005737(cytoplasm);GO:0005739(mitochondrion);GO:0043231(intracellular membrane-bounded organelle) | 00030(Pentose phosphate pathway) | 0.00 | 1 | 61, | 0 | -3.3000000000 | -2.4400000000 | 1.98 | 3' UTR | 1 | 9265530 | 9271337 | 5808 | 1 |
| chr1 | 9265104 | 9265313 | 210 | ENSG00000049239 | ENST00000495451 | -217 | H6PD | hexose-6-phosphate dehydrogenase/glucose 1-dehydrogenase [Source:HGNC Symbol;Acc:HGNC:4795] | GO:0016491(oxidoreductase activity);GO:0050661(NADP binding);GO:0055114(oxidation-reduction process);GO:0016787(hydrolase activity);GO:0005975(carbohydrate metabolic process);GO:0005783(endoplasmic reticulum);GO:0003824(catalytic activity);GO:0008152(metabolic process);GO:0005788(endoplasmic reticulum lumen);GO:0006006(glucose metabolic process);GO:0017057(6-phosphogluconolactonase activity);GO:0006098(pentose-phosphate shunt);GO:0004345(glucose-6-phosphate dehydrogenase activity);GO:0047936(glucose 1-dehydrogenase [NAD(P)] activity);GO:0030246(carbohydrate binding);GO:0006739(NADP metabolic process);GO:0097305(response to alcohol);GO:0005737(cytoplasm);GO:0005739(mitochondrion);GO:0043231(intracellular membrane-bounded organelle) | 00030(Pentose phosphate pathway) | 0.00 | 1 | 210, | 0 | -9.6000000000 | -8.5200000000 | 4.12 | 3' UTR | 1 | 9265530 | 9271337 | 5808 | 1 |
| chr1 | 9268940 | 9269269 | 330 | ENSG00000049239 | ENST00000495451 | 3410 | H6PD | hexose-6-phosphate dehydrogenase/glucose 1-dehydrogenase [Source:HGNC Symbol;Acc:HGNC:4795] | GO:0016491(oxidoreductase activity);GO:0050661(NADP binding);GO:0055114(oxidation-reduction process);GO:0016787(hydrolase activity);GO:0005975(carbohydrate metabolic process);GO:0005783(endoplasmic reticulum);GO:0003824(catalytic activity);GO:0008152(metabolic process);GO:0005788(endoplasmic reticulum lumen);GO:0006006(glucose metabolic process);GO:0017057(6-phosphogluconolactonase activity);GO:0006098(pentose-phosphate shunt);GO:0004345(glucose-6-phosphate dehydrogenase activity);GO:0047936(glucose 1-dehydrogenase [NAD(P)] activity);GO:0030246(carbohydrate binding);GO:0006739(NADP metabolic process);GO:0097305(response to alcohol);GO:0005737(cytoplasm);GO:0005739(mitochondrion);GO:0043231(intracellular membrane-bounded organelle) | 00030(Pentose phosphate pathway) | 0.00 | 1 | 330, | 0 | -47.2000000000 | -45.6000000000 | 9.69 | 3' UTR | 1 | 9265530 | 9271337 | 5808 | 1 |
| chr1 | 9269809 | 9269899 | 91 | ENSG00000049239 | ENST00000495451 | 4279 | H6PD | hexose-6-phosphate dehydrogenase/glucose 1-dehydrogenase [Source:HGNC Symbol;Acc:HGNC:4795] | GO:0016491(oxidoreductase activity);GO:0050661(NADP binding);GO:0055114(oxidation-reduction process);GO:0016787(hydrolase activity);GO:0005975(carbohydrate metabolic process);GO:0005783(endoplasmic reticulum);GO:0003824(catalytic activity);GO:0008152(metabolic process);GO:0005788(endoplasmic reticulum lumen);GO:0006006(glucose metabolic process);GO:0017057(6-phosphogluconolactonase activity);GO:0006098(pentose-phosphate shunt);GO:0004345(glucose-6-phosphate dehydrogenase activity);GO:0047936(glucose 1-dehydrogenase [NAD(P)] activity);GO:0030246(carbohydrate binding);GO:0006739(NADP metabolic process);GO:0097305(response to alcohol);GO:0005737(cytoplasm);GO:0005739(mitochondrion);GO:0043231(intracellular membrane-bounded organelle) | 00030(Pentose phosphate pathway) | 0.00 | 1 | 91, | 0 | -4.0100000000 | -3.1200000000 | 3.81 | 3' UTR | 1 | 9265530 | 9271337 | 5808 | 1 |
| chr1 | 9270648 | 9270887 | 240 | ENSG00000049239 | ENST00000495451 | 5118 | H6PD | hexose-6-phosphate dehydrogenase/glucose 1-dehydrogenase [Source:HGNC Symbol;Acc:HGNC:4795] | GO:0016491(oxidoreductase activity);GO:0050661(NADP binding);GO:0055114(oxidation-reduction process);GO:0016787(hydrolase activity);GO:0005975(carbohydrate metabolic process);GO:0005783(endoplasmic reticulum);GO:0003824(catalytic activity);GO:0008152(metabolic process);GO:0005788(endoplasmic reticulum lumen);GO:0006006(glucose metabolic process);GO:0017057(6-phosphogluconolactonase activity);GO:0006098(pentose-phosphate shunt);GO:0004345(glucose-6-phosphate dehydrogenase activity);GO:0047936(glucose 1-dehydrogenase [NAD(P)] activity);GO:0030246(carbohydrate binding);GO:0006739(NADP metabolic process);GO:0097305(response to alcohol);GO:0005737(cytoplasm);GO:0005739(mitochondrion);GO:0043231(intracellular membrane-bounded organelle) | 00030(Pentose phosphate pathway) | 0.00 | 1 | 240, | 0 | -6.1600000000 | -5.1700000000 | 3.08 | 3' UTR | 1 | 9265530 | 9271337 | 5808 | 1 |
| chr1 | 9355745 | 9356581 | 837 | ENSG00000171621 | ENST00000377399 | 38 | SPSB1 | splA/ryanodine receptor domain and SOCS box containing 1 [Source:HGNC Symbol;Acc:HGNC:30628] | GO:0005622(intracellular);GO:0005737(cytoplasm);GO:0016567(protein ubiquitination);GO:0035556(intracellular signal transduction);GO:0005515(protein binding);GO:0005829(cytosol);GO:0000209(protein polyubiquitination);GO:0043687(post-translational protein modification);GO:0004842(ubiquitin-protein transferase activity) | NA | 0.00 | 1 | 837, | 0 | -58.5000000000 | -56.8000000000 | 6.42 | 5' UTR | 1 | 9355707 | 9367575 | 11869 | 1 |
| chr1 | 9367652 | 9368338 | 687 | ENSG00000171621 | ENST00000377399 | 11945 | SPSB1 | splA/ryanodine receptor domain and SOCS box containing 1 [Source:HGNC Symbol;Acc:HGNC:30628] | GO:0005622(intracellular);GO:0005737(cytoplasm);GO:0016567(protein ubiquitination);GO:0035556(intracellular signal transduction);GO:0005515(protein binding);GO:0005829(cytosol);GO:0000209(protein polyubiquitination);GO:0043687(post-translational protein modification);GO:0004842(ubiquitin-protein transferase activity) | NA | 0.00 | 1 | 687, | 0 | -61.7000000000 | -60 | 15.9000000000 | 3' UTR | 1 | 9355707 | 9367575 | 11869 | 1 |
| chr1 | 9369263 | 9369472 | 210 | ENSG00000171621 | ENST00000377399 | 13556 | SPSB1 | splA/ryanodine receptor domain and SOCS box containing 1 [Source:HGNC Symbol;Acc:HGNC:30628] | GO:0005622(intracellular);GO:0005737(cytoplasm);GO:0016567(protein ubiquitination);GO:0035556(intracellular signal transduction);GO:0005515(protein binding);GO:0005829(cytosol);GO:0000209(protein polyubiquitination);GO:0043687(post-translational protein modification);GO:0004842(ubiquitin-protein transferase activity) | NA | 0.00 | 1 | 210, | 0 | -6.0900000000 | -5.1100000000 | 13.3000000000 | 3' UTR | 1 | 9355707 | 9367575 | 11869 | 1 |
| chr1 | 9602210 | 9602629 | 420 | ENSG00000188807 | ENST00000510900 | -3585 | TMEM201 | transmembrane protein 201 [Source:HGNC Symbol;Acc:HGNC:33719] | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005856(cytoskeleton);GO:0000922(spindle pole);GO:0005637(nuclear inner membrane);GO:0005521(lamin binding);GO:0051015(actin filament binding);GO:0007097(nuclear migration);GO:0010761(fibroblast migration);GO:0031965(nuclear membrane) | NA | 0.00 | 1 | 420, | 0 | -25.6000000000 | -24.3000000000 | 3.88 | 3' UTR | 1 | 9606214 | 9607685 | 1472 | 1 |
| chr1 | 9613259 | 9613438 | 180 | ENSG00000188807 | ENST00000508400 | 5531 | TMEM201 | transmembrane protein 201 [Source:HGNC Symbol;Acc:HGNC:33719] | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005856(cytoskeleton);GO:0000922(spindle pole);GO:0005637(nuclear inner membrane);GO:0005521(lamin binding);GO:0051015(actin filament binding);GO:0007097(nuclear migration);GO:0010761(fibroblast migration);GO:0031965(nuclear membrane) | NA | 0.00 | 1 | 180, | 0 | -5.9100000000 | -4.9400000000 | 2.70 | 3' UTR | 1 | 9607728 | 9613247 | 5520 | 1 |
| chr1 | 9726934 | 9727322 | 389 | ENSG00000171603 | ENST00000477264 | 6164 | CLSTN1 | calsyntenin 1 [Source:HGNC Symbol;Acc:HGNC:17447] | GO:0005634(nucleus);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005509(calcium ion binding);GO:0007155(cell adhesion);GO:0007156(homophilic cell adhesion via plasma membrane adhesion molecules);GO:0005794(Golgi apparatus);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0005886(plasma membrane);GO:0042995(cell projection);GO:0005515(protein binding);GO:0000139(Golgi membrane);GO:0030054(cell junction);GO:0045202(synapse);GO:0045211(postsynaptic membrane);GO:0070062(extracellular exosome);GO:0001540(amyloid-beta binding);GO:0019894(kinesin binding);GO:0042988(X11-like protein binding);GO:0001558(regulation of cell growth);GO:0050806(positive regulation of synaptic transmission);GO:0051965(positive regulation of synapse assembly);GO:0005576(extracellular region);GO:0005615(extracellular space);GO:0009986(cell surface);GO:0014069(postsynaptic density) | NA | 0.00 | 1 | 389, | 0 | -13.1000000000 | -11.9000000000 | 8.81 | 3' UTR | 1 | 9729028 | 9733486 | 4459 | 2 |
| chr1 | 9982658 | 9982807 | 150 | ENSG00000241326 | ENST00000413148 | -334 | AL603962.1 | NA | NA | NA | 0.00 | 1 | 150, | 0 | -3.8000000000 | -2.9100000000 | 4.29 | 3' UTR | 1 | 9983141 | 9984568 | 1428 | 1 |
| chr1 | 10033251 | 10033401 | 151 | ENSG00000130939 | ENST00000377153 | 0 | UBE4B | ubiquitination factor E4B [Source:HGNC Symbol;Acc:HGNC:12500] | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016740(transferase activity);GO:0016567(protein ubiquitination);GO:0004842(ubiquitin-protein transferase activity);GO:0006511(ubiquitin-dependent protein catabolic process);GO:0042787(protein ubiquitination involved in ubiquitin-dependent protein catabolic process);GO:0000151(ubiquitin ligase complex);GO:0034450(ubiquitin-ubiquitin ligase activity);GO:0000209(protein polyubiquitination);GO:0019899(enzyme binding);GO:0009411(response to UV);GO:0008626(granzyme-mediated apoptotic signaling pathway);GO:0030433(ubiquitin-dependent ERAD pathway);GO:0043161(proteasome-mediated ubiquitin-dependent protein catabolic process);GO:0005515(protein binding);GO:0005524(ATP binding);GO:0051117(ATPase binding);GO:0061630(ubiquitin protein ligase activity);GO:0003222(ventricular trabecula myocardium morphogenesis);GO:0006513(protein monoubiquitination);GO:0031175(neuron projection development);GO:0034976(response to endoplasmic reticulum stress);GO:0044257(cellular protein catabolic process);GO:0051865(protein autoubiquitination) | 04141(Protein processing in endoplasmic reticulum);04120(Ubiquitin mediated proteolysis) | 0.00 | 1 | 151, | 0 | -7.7400000000 | -6.7100000000 | 2.06 | 5' UTR | 1 | 10033274 | 10073118 | 39845 | 1 |
| chr1 | 10304074 | 10304554 | 481 | ENSG00000264501 | ENST00000584329 | -1911 | RN7SL731P | RNA, 7SL, cytoplasmic 731, pseudogene [Source:HGNC Symbol;Acc:HGNC:46747] | NA | NA | 0.00 | 1 | 481, | 0 | -8.8200000000 | -7.7600000000 | 2.77 | Exon (ENST00000377093/ENSG00000054523, exon 21 of 21) | 1 | 10306465 | 10306757 | 293 | 1 |
| chr1 | 10403109 | 10412872 | 9764 | ENSG00000142657 | ENST00000493288 | 0 | PGD | phosphogluconate dehydrogenase [Source:HGNC Symbol;Acc:HGNC:8891] | GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0004616(phosphogluconate dehydrogenase (decarboxylating) activity);GO:0006098(pentose-phosphate shunt);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0005829(cytosol);GO:0070062(extracellular exosome);GO:0009051(pentose-phosphate shunt, oxidative branch);GO:0019322(pentose biosynthetic process);GO:0019521(D-gluconate metabolic process) | 00030(Pentose phosphate pathway);00480(Glutathione metabolism) | 0.00 | 5 | 28,119,70,135,39, | 0,1052,4962,8309,9725 | -18 | -16.7000000000 | 60.9000000000 | 3' UTR | 1 | 10412834 | 10417044 | 4211 | 1 |
Definition of table:
| Term | Description |
|---|---|
| seqnames | The name of the chromosome (e.g. chr3, chrY, chr2 random) or scaffold (e.g. scaffold10671) |
| start | The starting position of the methylation site in the chromosome or scaffold. |
| end | The ending position of the RNA methylation site in the chromosome or scaffold. |
| width | Peak region of methylation |
| geneId | Defines the ID of gene on which the RNA methylation site locates |
| transcriptId | Defines the ID of transcript on which the RNA methylation site locates |
| distanceToTSS | Distrance from Peak site to transcription start site |
| geneName | Defines the name of gene (gene symbol) on which the RNA methylation site locates |
| GO | Gene ontology term and descriptionhttp://www.geneontology.org/ |
| KEGG | KEGG pathway annotation and description http://www.kegg.jp/kegg/kegg3a.html |
| score | p-value of the peak |
| blockCount | The number of blocks (exons) the RNA methylation site spans. |
| blockSizes | A comma-separated list of the block sizes. The number of items in this list should correspond to blockCount. |
| blockStarts | A comma-separated list of block starts. All of the blockStart positions should be calculated relative to chromStart. The number of items in this list should correspond to blockCount. |
| lg.p | log10(p-value) of the peak, indicating the significance of the peak as an RNA methylation site. |
| lg.fdr | log10(fdr) of the peak, indicating the significance of the peak as an RNA methylation site after multiple hypothesis correction. |
| fold_enrchment | fold enrichment within the peak in the IP sample compared with the input sample. |
| annotation | Location of peak, such as 3' UTR... |
| geneStart | The starting position of gene in the chromosome or scaffold. |
| geneEnd | The ending position of gene in the chromosome or scaffold. |
| geneLength | length of gene |
| geneStrand | 1=sense strand or "+"; 2=antisense strand or "-" |
document location: summary/3_peak_calling/*/*_peak.xlsx


document location:
summary/3_Peak_Calling/heatmap.png
summary/3_Peak_Calling/distProfile.png
| seqnames | start | end | width | geneId | transcriptId | distanceToTSS | geneName | Description | GO | KEGG | score | blockCount | blockSizes | blockStarts | lg.p | lg.fdr | fold_enrchment | diff.lg.fdr | diff.lg.p | diff.log2.fc | annotation | geneChr | geneStart | geneEnd | geneLength | geneStrand |
| chr1 | 1046342 | 1046449 | 108 | ENSG00000188157 | ENST00000466223 | -752 | AGRN | agrin [Source:HGNC Symbol;Acc:HGNC:329] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005829(cytosol);GO:0005509(calcium ion binding);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0007010(cytoskeleton organization);GO:0007165(signal transduction);GO:0005576(extracellular region);GO:0043236(laminin binding);GO:0007275(multicellular organism development);GO:0005578(proteinaceous extracellular matrix);GO:0030054(cell junction);GO:0045202(synapse);GO:0030154(cell differentiation);GO:0005200(structural constituent of cytoskeleton);GO:0007213(G-protein coupled acetylcholine receptor signaling pathway);GO:0043113(receptor clustering);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0070062(extracellular exosome);GO:0043547(positive regulation of GTPase activity);GO:0031012(extracellular matrix);GO:0043202(lysosomal lumen);GO:0002162(dystroglycan binding);GO:0033691(sialic acid binding);GO:0035374(chondroitin sulfate binding);GO:0043395(heparan sulfate proteoglycan binding);GO:0001523(retinoid metabolic process);GO:0006024(glycosaminoglycan biosynthetic process);GO:0006027(glycosaminoglycan catabolic process);GO:0030198(extracellular matrix organization);GO:0030203(glycosaminoglycan metabolic process);GO:0045162(clustering of voltage-gated sodium channels);GO:0045887(positive regulation of synaptic growth at neuromuscular junction);GO:0050808(synapse organization);GO:0051491(positive regulation of filopodium assembly);GO:0005605(basal lamina);GO:0005796(Golgi lumen) | 04512(ECM-receptor interaction) | 0.00 | 2 | 8,53, | 0,55 | -6.80 | -5.80 | 1.58 | -1.73 | -2.96 | -1.47 | Exon (ENST00000379370/ENSG00000188157, exon 18 of 36) | 1 | 1047201 | 1047865 | 665 | 1 |
| chr1 | 1048079 | 1048288 | 210 | ENSG00000188157 | ENST00000492947 | -241 | AGRN | agrin [Source:HGNC Symbol;Acc:HGNC:329] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005829(cytosol);GO:0005509(calcium ion binding);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0007010(cytoskeleton organization);GO:0007165(signal transduction);GO:0005576(extracellular region);GO:0043236(laminin binding);GO:0007275(multicellular organism development);GO:0005578(proteinaceous extracellular matrix);GO:0030054(cell junction);GO:0045202(synapse);GO:0030154(cell differentiation);GO:0005200(structural constituent of cytoskeleton);GO:0007213(G-protein coupled acetylcholine receptor signaling pathway);GO:0043113(receptor clustering);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0070062(extracellular exosome);GO:0043547(positive regulation of GTPase activity);GO:0031012(extracellular matrix);GO:0043202(lysosomal lumen);GO:0002162(dystroglycan binding);GO:0033691(sialic acid binding);GO:0035374(chondroitin sulfate binding);GO:0043395(heparan sulfate proteoglycan binding);GO:0001523(retinoid metabolic process);GO:0006024(glycosaminoglycan biosynthetic process);GO:0006027(glycosaminoglycan catabolic process);GO:0030198(extracellular matrix organization);GO:0030203(glycosaminoglycan metabolic process);GO:0045162(clustering of voltage-gated sodium channels);GO:0045887(positive regulation of synaptic growth at neuromuscular junction);GO:0050808(synapse organization);GO:0051491(positive regulation of filopodium assembly);GO:0005605(basal lamina);GO:0005796(Golgi lumen) | 04512(ECM-receptor interaction) | 0.00 | 1 | 210, | 0 | -28.10 | -26.80 | 1.77 | -1.56 | -2.74 | -0.44 | Exon (ENST00000379370/ENSG00000188157, exon 23 of 36) | 1 | 1048529 | 1049394 | 866 | 1 |
| chr1 | 1055429 | 1056118 | 690 | ENSG00000242590 | ENST00000418300 | 396 | AL645608.6 | NA | NA | NA | 0.00 | 1 | 690, | 0 | -192 | -190 | 8.10 | -1.37 | -2.47 | -0.24 | 3' UTR | 1 | 1055033 | 1056116 | 1084 | 1 |
| chr1 | 1327709 | 1328897 | 1189 | ENSG00000169962 | ENST00000339381 | -2417 | TAS1R3 | taste 1 receptor member 3 [Source:HGNC Symbol;Acc:HGNC:15661] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0004930(G-protein coupled receptor activity);GO:0007186(G-protein coupled receptor signaling pathway);GO:0005886(plasma membrane);GO:0004871(signal transducer activity);GO:0007165(signal transduction);GO:0050896(response to stimulus);GO:0050909(sensory perception of taste);GO:0046982(protein heterodimerization activity);GO:0008527(taste receptor activity);GO:0033041(sweet taste receptor activity);GO:0001582(detection of chemical stimulus involved in sensory perception of sweet taste);GO:0050916(sensory perception of sweet taste);GO:0050917(sensory perception of umami taste);GO:1903767(sweet taste receptor complex) | 04973(Carbohydrate digestion and absorption);04742(Taste transduction) | 0 | 1 | 1189, | 0 | -310 | -307 | 13.50 | -5.29 | -7.32 | -0.47 | 3' UTR | 1 | 1331314 | 1335306 | 3993 | 1 |
| chr1 | 1439706 | 1440904 | 1199 | ENSG00000179403 | ENST00000471398 | 3971 | VWA1 | von Willebrand factor A domain containing 1 [Source:HGNC Symbol;Acc:HGNC:30910] | GO:0005576(extracellular region);GO:0005578(proteinaceous extracellular matrix);GO:0005615(extracellular space);GO:0005604(basement membrane);GO:0070062(extracellular exosome);GO:0044267(cellular protein metabolic process);GO:0043687(post-translational protein modification);GO:0005788(endoplasmic reticulum lumen);GO:0042802(identical protein binding);GO:0030198(extracellular matrix organization);GO:0048266(behavioral response to pain);GO:0005614(interstitial matrix) | NA | 0 | 1 | 1199, | 0 | -1690 | -Inf | 14.20 | -5.65 | -7.74 | -0.35 | 3' UTR | 1 | 1435735 | 1437177 | 1443 | 1 |
| chr1 | 1534330 | 1534657 | 328 | ENSG00000205090 | ENST00000624426 | 5693 | TMEM240 | transmembrane protein 240 [Source:HGNC Symbol;Acc:HGNC:25186] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005886(plasma membrane);GO:0030054(cell junction);GO:0045202(synapse);GO:0097060(synaptic membrane) | NA | 0.00 | 1 | 328, | 0 | -18.10 | -16.90 | 4.18 | -2.70 | -4.22 | -1.67 | 3' UTR | 1 | 1539056 | 1540350 | 1295 | 2 |
| chr1 | 2563113 | 2563352 | 240 | ENSG00000157873 | ENST00000480305 | 1312 | TNFRSF14 | TNF receptor superfamily member 14 [Source:HGNC Symbol;Acc:HGNC:11912] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0002741(positive regulation of cytokine secretion involved in immune response);GO:0046642(negative regulation of alpha-beta T cell proliferation);GO:0050731(positive regulation of peptidyl-tyrosine phosphorylation);GO:0050829(defense response to Gram-negative bacterium);GO:0050830(defense response to Gram-positive bacterium);GO:2000406(positive regulation of T cell migration);GO:0009897(external side of plasma membrane);GO:0005515(protein binding);GO:0005886(plasma membrane);GO:0005887(integral component of plasma membrane);GO:0006954(inflammatory response);GO:0046718(viral entry into host cell);GO:0097190(apoptotic signaling pathway);GO:0033209(tumor necrosis factor-mediated signaling pathway);GO:0016032(viral process);GO:0031625(ubiquitin protein ligase binding);GO:0006955(immune response);GO:0007275(multicellular organism development);GO:0032496(response to lipopolysaccharide);GO:0042127(regulation of cell proliferation);GO:0007166(cell surface receptor signaling pathway);GO:0042981(regulation of apoptotic process);GO:0001618(virus receptor activity);GO:0031295(T cell costimulation);GO:0005031(tumor necrosis factor-activated receptor activity) | 04060(Cytokine-cytokine receptor interaction);05168(Herpes simplex infection) | 0.00 | 1 | 240, | 0 | -16 | -14.90 | 2.92 | -2.04 | -3.36 | -1.36 | 3' UTR | 1 | 2561801 | 2563452 | 1652 | 1 |
| chr1 | 2591051 | 2591469 | 419 | ENSG00000142606 | ENST00000471840 | 1209 | MMEL1 | membrane metalloendopeptidase like 1 [Source:HGNC Symbol;Acc:HGNC:14668] | GO:0006508(proteolysis);GO:0004222(metalloendopeptidase activity);GO:0008237(metallopeptidase activity);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0046872(metal ion binding);GO:0016787(hydrolase activity);GO:0008233(peptidase activity);GO:0005576(extracellular region) | NA | 0.00 | 1 | 419, | 0 | -289 | -286 | 47.50 | -2.13 | -3.48 | -0.61 | 3' UTR | 1 | 2590639 | 2592678 | 2040 | 2 |
| chr1 | 6613835 | 6614104 | 270 | ENSG00000116273 | ENST00000495385 | 73 | PHF13 | PHD finger protein 13 [Source:HGNC Symbol;Acc:HGNC:22983] | GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0016569(covalent chromatin modification);GO:0051301(cell division);GO:0007049(cell cycle);GO:0005654(nucleoplasm);GO:0007059(chromosome segregation);GO:0003682(chromatin binding);GO:0030261(chromosome condensation);GO:0000278(mitotic cell cycle);GO:0035064(methylated histone binding);GO:0007076(mitotic chromosome condensation) | NA | 0.00 | 1 | 270, | 0 | -21.90 | -20.70 | 4 | -1.53 | -2.69 | -1.34 | 5' UTR | 1 | 6613762 | 6622645 | 8884 | 1 |
| chr1 | 11023310 | 11023998 | 689 | ENSG00000120948 | ENST00000617757 | 0 | TARDBP | TAR DNA binding protein [Source:HGNC Symbol;Acc:HGNC:11571] | GO:0003676(nucleic acid binding);GO:0003723(RNA binding);GO:0005634(nucleus);GO:0003730(mRNA 3'-UTR binding);GO:0006397(mRNA processing);GO:0008380(RNA splicing);GO:0005654(nucleoplasm);GO:0006366(transcription from RNA polymerase II promoter);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0005515(protein binding);GO:0001205(transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding);GO:0032024(positive regulation of insulin secretion);GO:0034976(response to endoplasmic reticulum stress);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0005726(perichromatin fibrils);GO:0005737(cytoplasm);GO:0016607(nuclear speck);GO:0035061(interchromatin granule);GO:0042802(identical protein binding);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0003690(double-stranded DNA binding);GO:0070935(3'-UTR-mediated mRNA stabilization);GO:0010629(negative regulation of gene expression);GO:0051726(regulation of cell cycle);GO:0001933(negative regulation of protein phosphorylation);GO:0042981(regulation of apoptotic process);GO:0043922(negative regulation by host of viral transcription);GO:0071765(nuclear inner membrane organization) | NA | 0 | 1 | 689, | 0 | -327 | -Inf | 4.35 | -2.38 | -3.81 | 0.33 | 3' UTR | 1 | 11023323 | 11024152 | 830 | 1 |
| chr1 | 11750615 | 11754446 | 3832 | ENSG00000177674 | ENST00000471765 | 3103 | AGTRAP | angiotensin II receptor associated protein [Source:HGNC Symbol;Acc:HGNC:13539] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0043231(intracellular membrane-bounded organelle);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0005794(Golgi apparatus);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0005515(protein binding);GO:0000139(Golgi membrane);GO:0030659(cytoplasmic vesicle membrane);GO:0031410(cytoplasmic vesicle);GO:0038166(angiotensin-activated signaling pathway);GO:0004945(angiotensin type II receptor activity);GO:0001666(response to hypoxia);GO:0008217(regulation of blood pressure);GO:0005886(plasma membrane);GO:0005938(cell cortex) | NA | 0.00 | 2 | 157,22, | 0,3810 | -10.60 | -9.54 | 3.80 | -1.62 | -2.81 | -1.61 | 3' UTR | 1 | 11747512 | 11754802 | 7291 | 1 |
| chr1 | 12009708 | 12011957 | 2250 | ENSG00000270914 | ENST00000603287 | 5351 | AL096840.1 | NA | NA | NA | 0 | 2 | 19,462, | 0,1788 | -330 | -Inf | 5.02 | -1.55 | -2.72 | 0.30 | 3' UTR | 1 | 12017216 | 12017308 | 93 | 2 |
| chr1 | 15409895 | 15410163 | 269 | ENSG00000142634 | ENST00000375980 | 0 | EFHD2 | EF-hand domain family member D2 [Source:HGNC Symbol;Acc:HGNC:28670] | GO:0016020(membrane);GO:0046872(metal ion binding);GO:0005509(calcium ion binding);GO:0045121(membrane raft);GO:0045296(cadherin binding) | NA | 0.00 | 1 | 269, | 0 | -20.60 | -19.30 | 1.61 | -3.57 | -5.31 | -0.67 | 5' UTR | 1 | 15409895 | 15430343 | 20449 | 1 |
| chr1 | 15719789 | 15728093 | 8305 | ENSG00000116786 | ENST00000477849 | -1752 | PLEKHM2 | pleckstrin homology and RUN domain containing M2 [Source:HGNC Symbol;Acc:HGNC:29131] | GO:0005737(cytoplasm);GO:0005515(protein binding);GO:0019894(kinesin binding);GO:0007030(Golgi organization);GO:0032418(lysosome localization);GO:0032880(regulation of protein localization);GO:0010008(endosome membrane);GO:1903527(positive regulation of membrane tubulation) | 05132(Salmonella infection) | 0 | 5 | 132,60,229,819,15, | 0,1540,5528,7225,8290 | -524 | -Inf | 13.60 | -5.08 | -7.08 | -0.51 | Exon (ENST00000375799/ENSG00000116786, exon 6 of 20) | 1 | 15729845 | 15734115 | 4271 | 1 |
| chr1 | 15784595 | 15784893 | 299 | ENSG00000162458 | ENST00000509138 | 7522 | FBLIM1 | filamin binding LIM protein 1 [Source:HGNC Symbol;Acc:HGNC:24686] | GO:0001650(fibrillar center);GO:0005925(focal adhesion);GO:0030054(cell junction);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0007155(cell adhesion);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0005829(cytosol);GO:0001725(stress fiber);GO:0005938(cell cortex);GO:0008360(regulation of cell shape);GO:0016337(single organismal cell-cell adhesion);GO:0031005(filamin binding);GO:0033623(regulation of integrin activation);GO:0034329(cell junction assembly) | NA | 0.00 | 1 | 299, | 0 | -74.70 | -73.10 | 3 | -1.77 | -3.01 | -0.55 | 3' UTR | 1 | 15777073 | 15784838 | 7766 | 1 |
| chr1 | 19656869 | 19657463 | 595 | ENSG00000158748 | ENST00000289753 | -7824 | HTR6 | 5-hydroxytryptamine receptor 6 [Source:HGNC Symbol;Acc:HGNC:5301] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0004930(G-protein coupled receptor activity);GO:0007186(G-protein coupled receptor signaling pathway);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0004871(signal transducer activity);GO:0007165(signal transduction);GO:0004993(G-protein coupled serotonin receptor activity);GO:0098664(G-protein coupled serotonin receptor signaling pathway);GO:0005887(integral component of plasma membrane);GO:0007268(chemical synaptic transmission);GO:0032008(positive regulation of TOR signaling);GO:0030594(neurotransmitter receptor activity);GO:0007187(G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger);GO:0004969(histamine receptor activity);GO:0021795(cerebral cortex cell migration);GO:0005929(cilium) | 04020(Calcium signaling pathway);04024(cAMP signaling pathway);04080(Neuroactive ligand-receptor interaction);04726(Serotonergic synapse) | 0 | 1 | 595, | 0 | -1210 | -Inf | 30 | -2.29 | -3.70 | -0.30 | 3' UTR | 1 | 19665287 | 19679562 | 14276 | 1 |
| chr1 | 19656873 | 19657441 | 569 | ENSG00000158748 | ENST00000289753 | -7846 | HTR6 | 5-hydroxytryptamine receptor 6 [Source:HGNC Symbol;Acc:HGNC:5301] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0004930(G-protein coupled receptor activity);GO:0007186(G-protein coupled receptor signaling pathway);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0004871(signal transducer activity);GO:0007165(signal transduction);GO:0004993(G-protein coupled serotonin receptor activity);GO:0098664(G-protein coupled serotonin receptor signaling pathway);GO:0005887(integral component of plasma membrane);GO:0007268(chemical synaptic transmission);GO:0032008(positive regulation of TOR signaling);GO:0030594(neurotransmitter receptor activity);GO:0007187(G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger);GO:0004969(histamine receptor activity);GO:0021795(cerebral cortex cell migration);GO:0005929(cilium) | 04020(Calcium signaling pathway);04024(cAMP signaling pathway);04080(Neuroactive ligand-receptor interaction);04726(Serotonergic synapse) | 0 | 1 | 569, | 0 | -1210 | -Inf | 27.10 | -2.56 | -4.03 | -0.33 | 3' UTR | 1 | 19665287 | 19679562 | 14276 | 1 |
| chr1 | 21822198 | 21822707 | 510 | ENSG00000187942 | ENST00000484271 | 701 | LDLRAD2 | low density lipoprotein receptor class A domain containing 2 [Source:HGNC Symbol;Acc:HGNC:32071] | GO:0016020(membrane);GO:0016021(integral component of membrane) | NA | 0.00 | 1 | 510, | 0 | -103 | -101 | 24.50 | -1.45 | -2.59 | -0.64 | 3' UTR | 1 | 21821497 | 21822548 | 1052 | 1 |
| chr1 | 22864994 | 22865174 | 181 | ENSG00000133216 | ENST00000490436 | 0 | EPHB2 | EPH receptor B2 [Source:HGNC Symbol;Acc:HGNC:3393] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0000166(nucleotide binding);GO:0004672(protein kinase activity);GO:0005524(ATP binding);GO:0006468(protein phosphorylation);GO:0005634(nucleus);GO:0016740(transferase activity);GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0004713(protein tyrosine kinase activity);GO:0005003(ephrin receptor activity);GO:0007169(transmembrane receptor protein tyrosine kinase signaling pathway);GO:0005887(integral component of plasma membrane);GO:0005886(plasma membrane);GO:0042995(cell projection);GO:0005515(protein binding);GO:0004714(transmembrane receptor protein tyrosine kinase activity);GO:0007275(multicellular organism development);GO:0007399(nervous system development);GO:0030425(dendrite);GO:0005576(extracellular region);GO:0030424(axon);GO:0005829(cytosol);GO:0010628(positive regulation of gene expression);GO:0048013(ephrin receptor signaling pathway);GO:0060997(dendritic spine morphogenesis);GO:0060021(palate development);GO:0001525(angiogenesis);GO:0007411(axon guidance);GO:0071679(commissural neuron axon guidance);GO:0051965(positive regulation of synapse assembly);GO:0042472(inner ear morphogenesis);GO:0007612(learning);GO:0007611(learning or memory);GO:0018108(peptidyl-tyrosine phosphorylation);GO:0098794(postsynapse);GO:1900273(positive regulation of long-term synaptic potentiation);GO:0001540(amyloid-beta binding);GO:0005005(transmembrane-ephrin receptor activity);GO:0044877(macromolecular complex binding);GO:0001655(urogenital system development);GO:0001933(negative regulation of protein phosphorylation);GO:0007413(axonal fasciculation);GO:0022038(corpus callosum development);GO:0031915(positive regulation of synaptic plasticity);GO:0046580(negative regulation of Ras protein signal transduction);GO:0048168(regulation of neuronal synaptic plasticity);GO:0050878(regulation of body fluid levels);GO:0051389(inactivation of MAPKK activity);GO:0060996(dendritic spine development);GO:0070373(negative regulation of ERK1 and ERK2 cascade);GO:1903078(positive regulation of protein localization to plasma membrane);GO:1904782(negative regulation of NMDA glutamate receptor activity);GO:1904783(positive regulation of NMDA glutamate receptor activity);GO:0004872(receptor activity);GO:0005102(receptor binding);GO:0008046(axon guidance receptor activity);GO:0042802(identical protein binding);GO:0000902(cell morphogenesis);GO:0009887(animal organ morphogenesis);GO:0021631(optic nerve morphogenesis);GO:0021952(central nervous system projection neuron axonogenesis);GO:0031290(retinal ganglion cell axon guidance);GO:0048170(positive regulation of long-term neuronal synaptic plasticity);GO:0048593(camera-type eye morphogenesis);GO:0050770(regulation of axonogenesis);GO:0050771(negative regulation of axonogenesis);GO:0099557(trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission);GO:0106028;GO:0043025(neuronal cell body) | 04360(Axon guidance) | 0.00 | 1 | 181, | 0 | -5.34 | -4.39 | 1.32 | -1.52 | -2.68 | -0.66 | Exon (ENST00000544305/ENSG00000133216, exon 5 of 7) | 1 | 22865159 | 22882840 | 17682 | 1 |
| chr1 | 24469053 | 24469263 | 211 | ENSG00000001461 | ENST00000432012 | 12887 | NIPAL3 | NIPA like domain containing 3 [Source:HGNC Symbol;Acc:HGNC:25233] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005515(protein binding);GO:0015095(magnesium ion transmembrane transporter activity);GO:0015693(magnesium ion transport);GO:1903830(magnesium ion transmembrane transport) | NA | 0.00 | 1 | 211, | 0 | -6.64 | -5.65 | 2.20 | -2.10 | -3.44 | -1.81 | 3' UTR | 1 | 24456166 | 24466378 | 10213 | 1 |
| chr1 | 25498265 | 25498714 | 450 | ENSG00000157978 | ENST00000374338 | -44866 | LDLRAP1 | low density lipoprotein receptor adaptor protein 1 [Source:HGNC Symbol;Acc:HGNC:18640] | GO:0042632(cholesterol homeostasis);GO:0006810(transport);GO:0005737(cytoplasm);GO:0009967(positive regulation of signal transduction);GO:0006629(lipid metabolic process);GO:0005829(cytosol);GO:0008202(steroid metabolic process);GO:0005515(protein binding);GO:0008203(cholesterol metabolic process);GO:0005886(plasma membrane);GO:0006897(endocytosis);GO:0061024(membrane organization);GO:0005769(early endosome);GO:0009925(basal plasma membrane);GO:0048260(positive regulation of receptor-mediated endocytosis);GO:0009898(cytoplasmic side of plasma membrane);GO:0030424(axon);GO:0030665(clathrin-coated vesicle membrane);GO:0030159(receptor signaling complex scaffold activity);GO:0030276(clathrin binding);GO:0030121(AP-1 adaptor complex);GO:0035615(clathrin adaptor activity);GO:0030122(AP-2 adaptor complex);GO:0001540(amyloid-beta binding);GO:0034383(low-density lipoprotein particle clearance);GO:0090118(receptor-mediated endocytosis involved in cholesterol transport);GO:0055037(recycling endosome);GO:0050750(low-density lipoprotein particle receptor binding);GO:0006898(receptor-mediated endocytosis);GO:1905602(positive regulation of receptor-mediated endocytosis involved in cholesterol transport);GO:0001784(phosphotyrosine residue binding);GO:0090003(regulation of establishment of protein localization to plasma membrane);GO:0005883(neurofilament);GO:0005546(phosphatidylinositol-4,5-bisphosphate binding);GO:0031623(receptor internalization);GO:0043393(regulation of protein binding);GO:0035591(signaling adaptor activity);GO:0035612(AP-2 adaptor complex binding);GO:0042982(amyloid precursor protein metabolic process);GO:0090205(positive regulation of cholesterol metabolic process) | 04144(Endocytosis);04979(Cholesterol metabolism) | 0.00 | 1 | 450, | 0 | -15.30 | -14.10 | 4.33 | -1.76 | -3 | -1.04 | 3' UTR | 1 | 25543580 | 25568886 | 25307 | 1 |
| chr1 | 26169930 | 26170489 | 560 | ENSG00000142684 | ENST00000270812 | 0 | ZNF593 | zinc finger protein 593 [Source:HGNC Symbol;Acc:HGNC:30943] | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0008270(zinc ion binding);GO:0046872(metal ion binding);GO:0005730(nucleolus);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0005515(protein binding);GO:0030687(preribosome, large subunit precursor);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0043023(ribosomal large subunit binding);GO:0000055(ribosomal large subunit export from nucleus);GO:1903026(negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding) | NA | 0.00 | 2 | 254,72, | 0,488 | -117 | -115 | 4.95 | -3.29 | -4.97 | -0.83 | 5' UTR | 1 | 26169942 | 26170873 | 932 | 1 |
| chr1 | 26573328 | 26575030 | 1703 | ENSG00000117676 | ENST00000438977 | 1058 | RPS6KA1 | ribosomal protein S6 kinase A1 [Source:HGNC Symbol;Acc:HGNC:10430] | GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0000166(nucleotide binding);GO:0004672(protein kinase activity);GO:0004674(protein serine/threonine kinase activity);GO:0005524(ATP binding);GO:0006468(protein phosphorylation);GO:0005829(cytosol);GO:0016740(transferase activity);GO:0000287(magnesium ion binding);GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0035556(intracellular signal transduction);GO:0005515(protein binding);GO:0007049(cell cycle);GO:0005654(nucleoplasm);GO:0004712(protein serine/threonine/tyrosine kinase activity);GO:0043066(negative regulation of apoptotic process);GO:0007165(signal transduction);GO:0045597(positive regulation of cell differentiation);GO:0030307(positive regulation of cell growth);GO:0043154(negative regulation of cysteine-type endopeptidase activity involved in apoptotic process);GO:0043027(cysteine-type endopeptidase inhibitor activity involved in apoptotic process);GO:0043620(regulation of DNA-templated transcription in response to stress);GO:0006915(apoptotic process);GO:0072574(hepatocyte proliferation);GO:0043555(regulation of translation in response to stress);GO:2000491(positive regulation of hepatic stellate cell activation) | 04010(MAPK signaling pathway);04150(mTOR signaling pathway);04114(Oocyte meiosis);04914(Progesterone-mediated oocyte maturation);04720(Long-term potentiation);04722(Neurotrophin signaling pathway);04931(Insulin resistance) | 0.00 | 2 | 34,952, | 0,751 | -51.70 | -50.20 | 16.10 | -2.21 | -3.60 | -0.61 | 3' UTR | 1 | 26572270 | 26574734 | 2465 | 1 |
| chr1 | 26696040 | 26697419 | 1380 | ENSG00000117713 | ENST00000457599 | 0 | ARID1A | AT-rich interaction domain 1A [Source:HGNC Symbol;Acc:HGNC:11110] | GO:0005634(nucleus);GO:0005654(nucleoplasm);GO:0003677(DNA binding);GO:0006338(chromatin remodeling);GO:0090544(BAF-type complex);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0016569(covalent chromatin modification);GO:0005515(protein binding);GO:0007399(nervous system development);GO:0045893(positive regulation of transcription, DNA-templated);GO:0003713(transcription coactivator activity);GO:0000790(nuclear chromatin);GO:0030521(androgen receptor signaling pathway);GO:0016922(ligand-dependent nuclear receptor binding);GO:0031491(nucleosome binding);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0001704(formation of primary germ layer);GO:0001843(neural tube closure);GO:0003205(cardiac chamber development);GO:0003408(optic cup formation involved in camera-type eye development);GO:0006325(chromatin organization);GO:0006337(nucleosome disassembly);GO:0006344(maintenance of chromatin silencing);GO:0006357(regulation of transcription from RNA polymerase II promoter);GO:0007369(gastrulation);GO:0007566(embryo implantation);GO:0019827(stem cell population maintenance);GO:0030520(intracellular estrogen receptor signaling pathway);GO:0030900(forebrain development);GO:0042766(nucleosome mobilization);GO:0042921(glucocorticoid receptor signaling pathway);GO:0043044(ATP-dependent chromatin remodeling);GO:0048096(chromatin-mediated maintenance of transcription);GO:0055007(cardiac muscle cell differentiation);GO:0060674(placenta blood vessel development);GO:1901998(toxin transport);GO:0016514(SWI/SNF complex);GO:0071564(npBAF complex);GO:0071565(nBAF complex) | 05225(Hepatocellular carcinoma) | 0.00 | 1 | 1380, | 0 | -182 | -180 | 18.80 | -2.02 | -3.34 | -0.72 | 5' UTR | 1 | 26696404 | 26780817 | 84414 | 1 |
| chr1 | 26774760 | 26775000 | 241 | ENSG00000117713 | ENST00000532781 | 0 | ARID1A | AT-rich interaction domain 1A [Source:HGNC Symbol;Acc:HGNC:11110] | GO:0005634(nucleus);GO:0005654(nucleoplasm);GO:0003677(DNA binding);GO:0006338(chromatin remodeling);GO:0090544(BAF-type complex);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0016569(covalent chromatin modification);GO:0005515(protein binding);GO:0007399(nervous system development);GO:0045893(positive regulation of transcription, DNA-templated);GO:0003713(transcription coactivator activity);GO:0000790(nuclear chromatin);GO:0030521(androgen receptor signaling pathway);GO:0016922(ligand-dependent nuclear receptor binding);GO:0031491(nucleosome binding);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0001704(formation of primary germ layer);GO:0001843(neural tube closure);GO:0003205(cardiac chamber development);GO:0003408(optic cup formation involved in camera-type eye development);GO:0006325(chromatin organization);GO:0006337(nucleosome disassembly);GO:0006344(maintenance of chromatin silencing);GO:0006357(regulation of transcription from RNA polymerase II promoter);GO:0007369(gastrulation);GO:0007566(embryo implantation);GO:0019827(stem cell population maintenance);GO:0030520(intracellular estrogen receptor signaling pathway);GO:0030900(forebrain development);GO:0042766(nucleosome mobilization);GO:0042921(glucocorticoid receptor signaling pathway);GO:0043044(ATP-dependent chromatin remodeling);GO:0048096(chromatin-mediated maintenance of transcription);GO:0055007(cardiac muscle cell differentiation);GO:0060674(placenta blood vessel development);GO:1901998(toxin transport);GO:0016514(SWI/SNF complex);GO:0071564(npBAF complex);GO:0071565(nBAF complex) | 05225(Hepatocellular carcinoma) | 0.00 | 1 | 241, | 0 | -13.40 | -12.30 | 1.92 | -2.03 | -3.35 | -0.88 | 3' UTR | 1 | 26774870 | 26780983 | 6114 | 1 |
| chr1 | 26863228 | 26864457 | 1230 | ENSG00000175793 | ENST00000339276 | 90 | SFN | stratifin [Source:HGNC Symbol;Acc:HGNC:10773] | GO:0005739(mitochondrion);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0005829(cytosol);GO:0019904(protein domain specific binding);GO:0005515(protein binding);GO:0005576(extracellular region);GO:0071901(negative regulation of protein serine/threonine kinase activity);GO:0042802(identical protein binding);GO:0006977(DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest);GO:0045296(cadherin binding);GO:0070062(extracellular exosome);GO:0061024(membrane organization);GO:0007165(signal transduction);GO:0005615(extracellular space);GO:0061436(establishment of skin barrier);GO:1900740(positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway);GO:0030659(cytoplasmic vesicle membrane);GO:0001836(release of cytochrome c from mitochondria);GO:0043154(negative regulation of cysteine-type endopeptidase activity involved in apoptotic process);GO:0045606(positive regulation of epidermal cell differentiation);GO:0008426(protein kinase C inhibitor activity);GO:0019901(protein kinase binding);GO:0051219(phosphoprotein binding);GO:0000079(regulation of cyclin-dependent protein serine/threonine kinase activity);GO:0003334(keratinocyte development);GO:0006469(negative regulation of protein kinase activity);GO:0008630(intrinsic apoptotic signaling pathway in response to DNA damage);GO:0010482(regulation of epidermal cell division);GO:0010839(negative regulation of keratinocyte proliferation);GO:0030216(keratinocyte differentiation);GO:0030307(positive regulation of cell growth);GO:0031424(keratinization);GO:0043588(skin development);GO:0046827(positive regulation of protein export from nucleus);GO:0051726(regulation of cell cycle) | 04110(Cell cycle);04115(p53 signaling pathway);04960(Aldosterone-regulated sodium reabsorption) | 0 | 1 | 1230, | 0 | -570 | -Inf | 26.10 | -3.37 | -5.06 | -0.19 | 3' UTR | 1 | 26863138 | 26864457 | 1320 | 1 |
| chr1 | 26993707 | 26994096 | 390 | ENSG00000253368 | ENST00000522111 | 0 | TRNP1 | TMF1-regulated nuclear protein 1 [Source:HGNC Symbol;Acc:HGNC:34348] | GO:0005634(nucleus);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0007049(cell cycle);GO:0007275(multicellular organism development);GO:0007399(nervous system development);GO:0042127(regulation of cell proliferation);GO:0051726(regulation of cell cycle);GO:0005719(nuclear euchromatin);GO:0021696(cerebellar cortex morphogenesis);GO:0061351(neural precursor cell proliferation) | NA | 0.00 | 1 | 390, | 0 | -184 | -182 | 3.86 | -2.99 | -4.58 | -0.50 | 5' UTR | 1 | 26993707 | 27000898 | 7192 | 1 |
| chr1 | 26994545 | 27000120 | 5576 | ENSG00000253368 | ENST00000531285 | 226 | TRNP1 | TMF1-regulated nuclear protein 1 [Source:HGNC Symbol;Acc:HGNC:34348] | GO:0005634(nucleus);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0007049(cell cycle);GO:0007275(multicellular organism development);GO:0007399(nervous system development);GO:0042127(regulation of cell proliferation);GO:0051726(regulation of cell cycle);GO:0005719(nuclear euchromatin);GO:0021696(cerebellar cortex morphogenesis);GO:0061351(neural precursor cell proliferation) | NA | 0 | 2 | 86,274, | 0,5302 | -728 | -Inf | 3.37 | -12.10 | -14.90 | -0.43 | 3' UTR | 1 | 26994319 | 27000109 | 5791 | 1 |
| chr1 | 28742979 | 28743188 | 210 | ENSG00000198492 | ENST00000468863 | 3888 | YTHDF2 | YTH N6-methyladenosine RNA binding protein 2 [Source:HGNC Symbol;Acc:HGNC:31675] | GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0005829(cytosol);GO:0000932(P-body);GO:0005515(protein binding);GO:0006959(humoral immune response);GO:1990247(N6-methyladenosine-containing RNA binding);GO:0048598(embryonic morphogenesis);GO:0043488(regulation of mRNA stability);GO:0061157(mRNA destabilization);GO:1903679(positive regulation of cap-independent translational initiation) | NA | 0.00 | 1 | 210, | 0 | -10.90 | -9.80 | 1.68 | -1.32 | -2.41 | -0.63 | Exon (ENST00000542507/ENSG00000198492, exon 5 of 6) | 1 | 28739091 | 28742883 | 3793 | 1 |
| chr1 | 31348951 | 31364392 | 15442 | ENSG00000229044 | ENST00000430143 | -2152 | AL451070.1 | NA | NA | NA | 0.00 | 2 | 24,361, | 0,15081 | -48.60 | -47.10 | 3.74 | -1.72 | -2.94 | 0.57 | 3' UTR | 1 | 31333067 | 31346799 | 13733 | 2 |
| chr1 | 31434233 | 31434441 | 209 | ENSG00000168528 | ENST00000536384 | 20420 | SERINC2 | serine incorporator 2 [Source:HGNC Symbol;Acc:HGNC:23231] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0006665(sphingolipid metabolic process);GO:0006658(phosphatidylserine metabolic process);GO:0070062(extracellular exosome);GO:0015194(L-serine transmembrane transporter activity);GO:0015825(L-serine transport);GO:1904219(positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity);GO:1904222(positive regulation of serine C-palmitoyltransferase activity) | NA | 0.00 | 1 | 209, | 0 | -20.60 | -19.40 | 2.11 | -2.64 | -4.15 | -0.88 | 3' UTR | 1 | 31413813 | 31434680 | 20868 | 1 |
| chr1 | 31587030 | 31587626 | 597 | ENSG00000142910 | ENST00000480586 | 1879 | TINAGL1 | tubulointerstitial nephritis antigen like 1 [Source:HGNC Symbol;Acc:HGNC:19168] | GO:0005615(extracellular space);GO:0005515(protein binding);GO:0008234(cysteine-type peptidase activity);GO:0006508(proteolysis);GO:0005576(extracellular region);GO:0006955(immune response);GO:0005044(scavenger receptor activity);GO:0006898(receptor-mediated endocytosis);GO:0030247(polysaccharide binding);GO:0070062(extracellular exosome);GO:0007155(cell adhesion);GO:0016197(endosomal transport);GO:0031012(extracellular matrix);GO:0005201(extracellular matrix structural constituent);GO:0043236(laminin binding);GO:0005737(cytoplasm) | NA | 0.00 | 1 | 597, | 0 | -85.30 | -83.70 | 3.02 | -2.18 | -3.56 | -0.37 | 3' UTR | 1 | 31585151 | 31586442 | 1292 | 1 |
| chr1 | 32193259 | 32195421 | 2163 | ENSG00000160050 | ENST00000373602 | -4965 | CCDC28B | coiled-coil domain containing 28B [Source:HGNC Symbol;Acc:HGNC:28163] | GO:0005737(cytoplasm);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0005813(centrosome);GO:0005815(microtubule organizing center);GO:0060271(cilium assembly);GO:0030030(cell projection organization) | NA | 0.00 | 3 | 42,96,520, | 0,806,1643 | -129 | -128 | 9.21 | -1.38 | -2.49 | -0.29 | 3' UTR | 1 | 32200386 | 32205387 | 5002 | 1 |
| chr1 | 32770981 | 32771550 | 570 | ENSG00000162522 | ENST00000373480 | 5314 | KIAA1522 | KIAA1522 [Source:HGNC Symbol;Acc:HGNC:29301] | GO:0030154(cell differentiation) | NA | 0.00 | 1 | 570, | 0 | -296 | -293 | 3.67 | -3.38 | -5.07 | -0.31 | Exon (ENST00000401073/ENSG00000162522, exon 6 of 7) | 1 | 32765667 | 32774970 | 9304 | 1 |
| chr1 | 33281593 | 33281831 | 239 | ENSG00000160094 | ENST00000477934 | 1243 | ZNF362 | zinc finger protein 362 [Source:HGNC Symbol;Acc:HGNC:18079] | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding) | NA | 0.00 | 1 | 239, | 0 | -37.40 | -36 | 4.54 | -1.61 | -2.80 | 1.09 | Exon (ENST00000539719/ENSG00000160094, exon 6 of 9) | 1 | 33280350 | 33281973 | 1624 | 1 |
| chr1 | 36156199 | 36171546 | 15348 | ENSG00000116871 | ENST00000373151 | 0 | MAP7D1 | MAP7 domain containing 1 [Source:HGNC Symbol;Acc:HGNC:25514] | GO:0000226(microtubule cytoskeleton organization);GO:0015630(microtubule cytoskeleton);GO:0005737(cytoplasm);GO:0005829(cytosol);GO:0005856(cytoskeleton);GO:0005819(spindle);GO:0005198(structural molecule activity) | NA | 0 | 4 | 265,76,345,34, | 0,1016,14772,15314 | -938 | -Inf | 8.10 | -2.99 | -4.58 | -0.27 | 5' UTR | 1 | 36156202 | 36180840 | 24639 | 1 |
| chr1 | 36180310 | 36180849 | 540 | ENSG00000116871 | ENST00000487114 | 1289 | MAP7D1 | MAP7 domain containing 1 [Source:HGNC Symbol;Acc:HGNC:25514] | GO:0000226(microtubule cytoskeleton organization);GO:0015630(microtubule cytoskeleton);GO:0005737(cytoplasm);GO:0005829(cytosol);GO:0005856(cytoskeleton);GO:0005819(spindle);GO:0005198(structural molecule activity) | NA | 0.00 | 1 | 540, | 0 | -175 | -173 | 22.90 | -1.31 | -2.39 | -0.27 | 3' UTR | 1 | 36179021 | 36180315 | 1295 | 1 |
| chr1 | 36296667 | 36304308 | 7642 | ENSG00000214193 | ENST00000453908 | -2079 | SH3D21 | SH3 domain containing 21 [Source:HGNC Symbol;Acc:HGNC:26236] | GO:0005634(nucleus);GO:0005654(nucleoplasm);GO:0005886(plasma membrane);GO:0070062(extracellular exosome) | NA | 0 | 4 | 104,199,144,513, | 0,4219,4886,7129 | -516 | -Inf | 23.60 | -3.44 | -5.15 | 0.48 | 3' UTR | 1 | 36306387 | 36321230 | 14844 | 1 |
| chr1 | 36322379 | 36322529 | 151 | ENSG00000142694 | ENST00000490466 | 1086 | EVA1B | eva-1 homolog B [Source:HGNC Symbol;Acc:HGNC:25558] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005515(protein binding) | NA | 0.00 | 1 | 151, | 0 | -16.50 | -15.30 | 2.15 | -2.08 | -3.42 | -0.80 | 3' UTR | 1 | 36322281 | 36323615 | 1335 | 2 |
| chr1 | 37799584 | 37800211 | 628 | ENSG00000233728 | ENST00000433474 | 668 | AL929472.3 | NA | NA | NA | 0.00 | 1 | 628, | 0 | -246 | -244 | 3.73 | -1.68 | -2.88 | -0.29 | 3' UTR | 1 | 37799720 | 37800879 | 1160 | 2 |
| chr1 | 37808109 | 37808495 | 387 | ENSG00000196449 | ENST00000373044 | 0 | YRDC | yrdC N6-threonylcarbamoyltransferase domain containing [Source:HGNC Symbol;Acc:HGNC:28905] | GO:0016020(membrane);GO:0005739(mitochondrion);GO:0003725(double-stranded RNA binding);GO:0005737(cytoplasm);GO:0000049(tRNA binding);GO:0051051(negative regulation of transport);GO:0016779(nucleotidyltransferase activity);GO:0002949(tRNA threonylcarbamoyladenosine modification);GO:0006450(regulation of translational fidelity) | NA | 0.00 | 1 | 387, | 0 | -121 | -120 | 11.60 | -5.53 | -7.61 | -1.11 | 5' UTR | 1 | 37802944 | 37808185 | 5242 | 2 |
| chr1 | 39026376 | 39034636 | 8261 | ENSG00000168653 | ENST00000372969 | 58 | NDUFS5 | NADH:ubiquinone oxidoreductase subunit S5 [Source:HGNC Symbol;Acc:HGNC:7712] | GO:0016020(membrane);GO:0055114(oxidation-reduction process);GO:0005743(mitochondrial inner membrane);GO:0005739(mitochondrion);GO:0070469(respiratory chain);GO:0005758(mitochondrial intermembrane space);GO:0032981(mitochondrial respiratory chain complex I assembly);GO:0008137(NADH dehydrogenase (ubiquinone) activity);GO:0006120(mitochondrial electron transport, NADH to ubiquinone);GO:0005747(mitochondrial respiratory chain complex I) | 00190(Oxidative phosphorylation);04723(Retrograde endocannabinoid signaling);05010(Alzheimer's disease);05012(Parkinson's disease);05016(Huntington's disease);04932(Non-alcoholic fatty liver disease (NAFLD)) | 0 | 3 | 27,218,245, | 0,2347,8016 | -353 | -Inf | 113 | -1.40 | -2.51 | -0.80 | 5' UTR | 1 | 39026318 | 39034636 | 8319 | 1 |
| chr1 | 39388387 | 39409531 | 21145 | ENSG00000127603 | ENST00000530275 | 0 | MACF1 | microtubule-actin crosslinking factor 1 [Source:HGNC Symbol;Acc:HGNC:13664] | GO:0003779(actin binding);GO:0005509(calcium ion binding);GO:0005856(cytoskeleton);GO:0008017(microtubule binding);GO:0016055(Wnt signaling pathway);GO:0016887(ATPase activity);GO:0003723(RNA binding);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0046872(metal ion binding);GO:0005794(Golgi apparatus);GO:0005886(plasma membrane);GO:0042995(cell projection);GO:0005515(protein binding);GO:0005874(microtubule);GO:0045296(cadherin binding);GO:0051015(actin filament binding);GO:0045773(positive regulation of axon extension);GO:0032587(ruffle membrane);GO:0042060(wound healing);GO:0051893(regulation of focal adhesion assembly);GO:0032886(regulation of microtubule-based process);GO:0030177(positive regulation of Wnt signaling pathway);GO:0010632(regulation of epithelial cell migration);GO:0043001(Golgi to plasma membrane protein transport);GO:0016021(integral component of membrane);GO:0030054(cell junction);GO:0015629(actin cytoskeleton) | NA | 0.00 | 2 | 272,29, | 0,21116 | -94.90 | -93.20 | 2.87 | -2.41 | -3.85 | 0.53 | 5' UTR | 1 | 39409503 | 39428092 | 18590 | 1 |
| chr1 | 39412590 | 39412830 | 241 | ENSG00000127603 | ENST00000641104 | 1567 | MACF1 | microtubule-actin crosslinking factor 1 [Source:HGNC Symbol;Acc:HGNC:13664] | GO:0003779(actin binding);GO:0005509(calcium ion binding);GO:0005856(cytoskeleton);GO:0008017(microtubule binding);GO:0016055(Wnt signaling pathway);GO:0016887(ATPase activity);GO:0003723(RNA binding);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0046872(metal ion binding);GO:0005794(Golgi apparatus);GO:0005886(plasma membrane);GO:0042995(cell projection);GO:0005515(protein binding);GO:0005874(microtubule);GO:0045296(cadherin binding);GO:0051015(actin filament binding);GO:0045773(positive regulation of axon extension);GO:0032587(ruffle membrane);GO:0042060(wound healing);GO:0051893(regulation of focal adhesion assembly);GO:0032886(regulation of microtubule-based process);GO:0030177(positive regulation of Wnt signaling pathway);GO:0010632(regulation of epithelial cell migration);GO:0043001(Golgi to plasma membrane protein transport);GO:0016021(integral component of membrane);GO:0030054(cell junction);GO:0015629(actin cytoskeleton) | NA | 0.00 | 1 | 241, | 0 | -12.30 | -11.20 | 1.41 | -1.77 | -3.01 | 0.42 | Exon (ENST00000530275/ENSG00000127603, exon 1 of 6) | 1 | 39411023 | 39416482 | 5460 | 1 |
| chr1 | 39485708 | 39485888 | 181 | ENSG00000183682 | ENST00000331593 | -5758 | BMP8A | bone morphogenetic protein 8a [Source:HGNC Symbol;Acc:HGNC:21650] | GO:0008083(growth factor activity);GO:0005576(extracellular region);GO:0007275(multicellular organism development);GO:0005125(cytokine activity);GO:0005615(extracellular space);GO:0030154(cell differentiation);GO:0060395(SMAD protein signal transduction);GO:0001503(ossification);GO:0051216(cartilage development);GO:0005160(transforming growth factor beta receptor binding);GO:0010862(positive regulation of pathway-restricted SMAD protein phosphorylation);GO:0030509(BMP signaling pathway);GO:0042981(regulation of apoptotic process);GO:0043408(regulation of MAPK cascade);GO:0048468(cell development);GO:2000505(regulation of energy homeostasis);GO:0046676(negative regulation of insulin secretion);GO:0070700(BMP receptor binding);GO:0002024(diet induced thermogenesis) | 04350(TGF-beta signaling pathway);04390(Hippo signaling pathway) | 0.00 | 1 | 181, | 0 | -43.40 | -41.90 | 1.84 | -3.02 | -4.62 | 0.44 | 3' UTR | 1 | 39491646 | 39525935 | 34290 | 1 |
| chr1 | 43359081 | 43359533 | 453 | ENSG00000117399 | ENST00000372462 | 68 | CDC20 | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | GO:0005737(cytoplasm);GO:0016567(protein ubiquitination);GO:0051301(cell division);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0007049(cell cycle);GO:0007399(nervous system development);GO:0030154(cell differentiation);GO:0031145(anaphase-promoting complex-dependent catabolic process);GO:0000922(spindle pole);GO:0005815(microtubule organizing center);GO:0005654(nucleoplasm);GO:0005829(cytosol);GO:0016579(protein deubiquitination);GO:0043161(proteasome-mediated ubiquitin-dependent protein catabolic process);GO:0051437(positive regulation of ubiquitin-protein ligase activity involved in regulation of mitotic cell cycle transition);GO:0008022(protein C-terminus binding);GO:0010997(anaphase-promoting complex binding);GO:0097027(ubiquitin-protein transferase activator activity);GO:1904668(positive regulation of ubiquitin protein ligase activity);GO:0007062(sister chromatid cohesion);GO:0019899(enzyme binding);GO:0051436(negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle);GO:0042787(protein ubiquitination involved in ubiquitin-dependent protein catabolic process);GO:0051439(regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle);GO:0005680(anaphase-promoting complex);GO:0005819(spindle);GO:0090129(positive regulation of synapse maturation);GO:0031915(positive regulation of synaptic plasticity);GO:0042826(histone deacetylase binding);GO:0007064(mitotic sister chromatid cohesion);GO:0008284(positive regulation of cell proliferation);GO:0040020(regulation of meiotic nuclear division);GO:0050773(regulation of dendrite development);GO:0090307(mitotic spindle assembly);GO:0005813(centrosome);GO:0043234(protein complex);GO:0048471(perinuclear region of cytoplasm) | 04120(Ubiquitin mediated proteolysis);04110(Cell cycle);04114(Oocyte meiosis);05203(Viral carcinogenesis);05166(HTLV-I infection) | 0 | 2 | 316,44, | 0,409 | -434 | -Inf | 8.93 | -2.16 | -3.52 | -0.57 | 5' UTR | 1 | 43359013 | 43363203 | 4191 | 1 |
| chr1 | 43603627 | 43604166 | 540 | ENSG00000142949 | ENST00000463041 | -1416 | PTPRF | protein tyrosine phosphatase, receptor type F [Source:HGNC Symbol;Acc:HGNC:9670] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0016787(hydrolase activity);GO:0007155(cell adhesion);GO:0004725(protein tyrosine phosphatase activity);GO:0016791(phosphatase activity);GO:0006470(protein dephosphorylation);GO:0016311(dephosphorylation);GO:0004721(phosphoprotein phosphatase activity);GO:0005886(plasma membrane);GO:0008201(heparin binding);GO:0070062(extracellular exosome);GO:0035335(peptidyl-tyrosine dephosphorylation);GO:0005887(integral component of plasma membrane);GO:0016477(cell migration);GO:0007185(transmembrane receptor protein tyrosine phosphatase signaling pathway);GO:0005001(transmembrane receptor protein tyrosine phosphatase activity);GO:0032403(protein complex binding);GO:0035373(chondroitin sulfate proteoglycan binding);GO:0010975(regulation of neuron projection development);GO:0031102(neuron projection regeneration);GO:0048679(regulation of axon regeneration);GO:1900121(negative regulation of receptor binding);GO:0043005(neuron projection);GO:0043025(neuronal cell body) | 04514(Cell adhesion molecules (CAMs));04520(Adherens junction);04910(Insulin signaling pathway);04931(Insulin resistance) | 0 | 1 | 540, | 0 | -1250 | -Inf | 5.59 | -5.19 | -7.20 | -0.30 | Exon (ENST00000359947/ENSG00000142949, exon 16 of 34) | 1 | 43605582 | 43606868 | 1287 | 1 |
| chr1 | 43967603 | 43968022 | 420 | ENSG00000117408 | ENST00000486876 | 443 | IPO13 | importin 13 [Source:HGNC Symbol;Acc:HGNC:16853] | GO:0006810(transport);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0006886(intracellular protein transport);GO:0015031(protein transport);GO:0005515(protein binding);GO:0008536(Ran GTPase binding);GO:0008139(nuclear localization sequence binding);GO:0008565(protein transporter activity);GO:0031965(nuclear membrane);GO:0006606(protein import into nucleus) | NA | 0.00 | 1 | 420, | 0 | -11.90 | -10.80 | 8.32 | -1.36 | -2.45 | -1.48 | 3' UTR | 1 | 43967160 | 43967663 | 504 | 1 |
| chr1 | 43972303 | 43972592 | 290 | ENSG00000132768 | ENST00000527319 | 834 | DPH2 | DPH2 homolog [Source:HGNC Symbol;Acc:HGNC:3004] | GO:0016740(transferase activity);GO:0005515(protein binding);GO:0005829(cytosol);GO:0017183(peptidyl-diphthamide biosynthetic process from peptidyl-histidine);GO:0090560(2-(3-amino-3-carboxypropyl)histidine synthase activity) | NA | 0.00 | 2 | 32,178, | 0,112 | -36.90 | -35.60 | 2.55 | -1.35 | -2.45 | -0.54 | 3' UTR | 1 | 43971469 | 43972808 | 1340 | 1 |
| chr1 | 43990362 | 43990781 | 420 | ENSG00000159214 | ENST00000486064 | -578 | CCDC24 | coiled-coil domain containing 24 [Source:HGNC Symbol;Acc:HGNC:28688] | GO:0005515(protein binding) | NA | 0.00 | 1 | 420, | 0 | -144 | -142 | 2.31 | -1.47 | -2.62 | -0.23 | 3' UTR | 1 | 43991359 | 43996524 | 5166 | 1 |
| chr1 | 44632034 | 44632296 | 263 | ENSG00000187147 | ENST00000497469 | 0 | RNF220 | ring finger protein 220 [Source:HGNC Symbol;Acc:HGNC:25552] | GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0016740(transferase activity);GO:0016567(protein ubiquitination);GO:0005515(protein binding);GO:0090263(positive regulation of canonical Wnt signaling pathway);GO:0004842(ubiquitin-protein transferase activity);GO:0051865(protein autoubiquitination);GO:0061630(ubiquitin protein ligase activity) | NA | 0.00 | 2 | 179,62, | 0,201 | -49.80 | -48.30 | 16.10 | -3.07 | -4.68 | -1.08 | 5' UTR | 1 | 44632235 | 44645081 | 12847 | 1 |
| chr1 | 45508955 | 45509194 | 240 | ENSG00000132763 | ENST00000477188 | -239 | MMACHC | methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria [Source:HGNC Symbol;Acc:HGNC:24525] | GO:0005737(cytoplasm);GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0042803(protein homodimerization activity);GO:0005515(protein binding);GO:0005829(cytosol);GO:0031419(cobalamin binding);GO:0071949(FAD binding);GO:0009235(cobalamin metabolic process);GO:0043295(glutathione binding);GO:0006749(glutathione metabolic process);GO:0032451(demethylase activity);GO:0009236(cobalamin biosynthetic process);GO:0033787(cyanocobalamin reductase (cyanide-eliminating) activity);GO:0070988(demethylation);GO:0005739(mitochondrion) | 04977(Vitamin digestion and absorption) | 0.00 | 1 | 240, | 0 | -28.60 | -27.30 | 2.56 | -1.46 | -2.60 | -0.70 | Exon (ENST00000401061/ENSG00000132763, exon 4 of 4) | 1 | 45509433 | 45510505 | 1073 | 1 |
| chr1 | 45606656 | 45607836 | 1181 | ENSG00000281825 | ENST00000626823 | -926 | AL355480.4 | NA | NA | NA | 0 | 2 | 25,516, | 0,665 | -331 | -Inf | 6.35 | -3.85 | -5.64 | 0.52 | 3' UTR | 1 | 45605657 | 45605730 | 74 | 2 |
| chr1 | 50970312 | 50974304 | 3993 | ENSG00000123080 | ENST00000371761 | 368 | CDKN2C | cyclin dependent kinase inhibitor 2C [Source:HGNC Symbol;Acc:HGNC:1789] | GO:0008285(negative regulation of cell proliferation);GO:0005737(cytoplasm);GO:0005634(nucleus);GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0005515(protein binding);GO:0007049(cell cycle);GO:0005829(cytosol);GO:0019901(protein kinase binding);GO:0030308(negative regulation of cell growth);GO:0007050(cell cycle arrest);GO:0000082(G1/S transition of mitotic cell cycle);GO:0004861(cyclin-dependent protein serine/threonine kinase inhibitor activity);GO:0042326(negative regulation of phosphorylation);GO:0000079(regulation of cyclin-dependent protein serine/threonine kinase activity);GO:0045736(negative regulation of cyclin-dependent protein serine/threonine kinase activity);GO:0048709(oligodendrocyte differentiation) | 04110(Cell cycle);05202(Transcriptional misregulation in cancers);05166(HTLV-I infection);01522(Endocrine resistance) | 0.00 | 2 | 186,412, | 0,3581 | -131 | -129 | 4.45 | -1.56 | -2.74 | -0.48 | 5' UTR | 1 | 50969944 | 50974558 | 4615 | 1 |
| chr1 | 55064345 | 55064673 | 329 | ENSG00000162402 | ENST00000480962 | 8319 | USP24 | ubiquitin specific peptidase 24 [Source:HGNC Symbol;Acc:HGNC:12623] | GO:0016579(protein deubiquitination);GO:0016787(hydrolase activity);GO:0008233(peptidase activity);GO:0008234(cysteine-type peptidase activity);GO:0006508(proteolysis);GO:0006511(ubiquitin-dependent protein catabolic process);GO:0036459(thiol-dependent ubiquitinyl hydrolase activity);GO:0005654(nucleoplasm) | NA | 0.00 | 1 | 329, | 0 | -8.58 | -7.54 | 1.83 | -2.31 | -3.72 | -0.84 | 3' UTR | 1 | 55071813 | 55072992 | 1180 | 2 |
| chr1 | 85582855 | 85583304 | 450 | ENSG00000142871 | ENST00000480413 | 1655 | CYR61 | cysteine rich angiogenic inducer 61 [Source:HGNC Symbol;Acc:HGNC:2654] | GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0001934(positive regulation of protein phosphorylation);GO:0030335(positive regulation of cell migration);GO:0030513(positive regulation of BMP signaling pathway);GO:0070372(regulation of ERK1 and ERK2 cascade);GO:0000166(nucleotide binding);GO:0007155(cell adhesion);GO:0005576(extracellular region);GO:0005520(insulin-like growth factor binding);GO:0001558(regulation of cell growth);GO:0006935(chemotaxis);GO:0008201(heparin binding);GO:0044267(cellular protein metabolic process);GO:0031012(extracellular matrix);GO:0043687(post-translational protein modification);GO:0007267(cell-cell signaling);GO:0005578(proteinaceous extracellular matrix);GO:0045669(positive regulation of osteoblast differentiation);GO:0005788(endoplasmic reticulum lumen);GO:0009653(anatomical structure morphogenesis);GO:0008283(cell proliferation);GO:0045860(positive regulation of protein kinase activity);GO:0005178(integrin binding);GO:0019838(growth factor binding);GO:0050840(extracellular matrix binding);GO:0001649(osteoblast differentiation);GO:0002041(intussusceptive angiogenesis);GO:0003181(atrioventricular valve morphogenesis);GO:0003278(apoptotic process involved in heart morphogenesis);GO:0003281(ventricular septum development);GO:0010518(positive regulation of phospholipase activity);GO:0010811(positive regulation of cell-substrate adhesion);GO:0016337(single organismal cell-cell adhesion);GO:0030198(extracellular matrix organization);GO:0033690(positive regulation of osteoblast proliferation);GO:0043065(positive regulation of apoptotic process);GO:0043066(negative regulation of apoptotic process);GO:0043280(positive regulation of cysteine-type endopeptidase activity involved in apoptotic process);GO:0044319(wound healing, spreading of cells);GO:0045597(positive regulation of cell differentiation);GO:0060413(atrial septum morphogenesis);GO:0060548(negative regulation of cell death);GO:0060591(chondroblast differentiation);GO:0060710(chorio-allantoic fusion);GO:0060716(labyrinthine layer blood vessel development);GO:0061036(positive regulation of cartilage development);GO:0072593(reactive oxygen species metabolic process);GO:2000304(positive regulation of ceramide biosynthetic process);GO:0005615(extracellular space) | NA | 0 | 1 | 450, | 0 | -661 | -Inf | 5.26 | -1.35 | -2.45 | 0.17 | 3' UTR | 1 | 85581200 | 85582099 | 900 | 1 |
| chr1 | 109929406 | 109930004 | 599 | ENSG00000184371 | ENST00000526001 | 13749 | CSF1 | colony stimulating factor 1 [Source:HGNC Symbol;Acc:HGNC:2432] | GO:0016021(integral component of membrane);GO:0008083(growth factor activity);GO:0005125(cytokine activity);GO:0005615(extracellular space);GO:0010628(positive regulation of gene expression);GO:0048471(perinuclear region of cytoplasm);GO:0030335(positive regulation of cell migration);GO:0016020(membrane);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0005576(extracellular region);GO:0030154(cell differentiation);GO:0002376(immune system process);GO:0045087(innate immune response);GO:0006954(inflammatory response);GO:0070062(extracellular exosome);GO:0044267(cellular protein metabolic process);GO:0043687(post-translational protein modification);GO:0008283(cell proliferation);GO:0008284(positive regulation of cell proliferation);GO:0042803(protein homodimerization activity);GO:0005788(endoplasmic reticulum lumen);GO:0030097(hemopoiesis);GO:0001954(positive regulation of cell-matrix adhesion);GO:0045860(positive regulation of protein kinase activity);GO:0042117(monocyte activation);GO:0030316(osteoclast differentiation);GO:0045672(positive regulation of osteoclast differentiation);GO:0010744(positive regulation of macrophage derived foam cell differentiation);GO:0032270(positive regulation of cellular protein metabolic process);GO:0045651(positive regulation of macrophage differentiation);GO:0007169(transmembrane receptor protein tyrosine kinase signaling pathway);GO:0030225(macrophage differentiation);GO:1990682(CSF1-CSF1R complex);GO:0010759(positive regulation of macrophage chemotaxis);GO:0005157(macrophage colony-stimulating factor receptor binding);GO:0002158(osteoclast proliferation);GO:0003006(developmental process involved in reproduction);GO:0010743(regulation of macrophage derived foam cell differentiation);GO:0030278(regulation of ossification);GO:0032946(positive regulation of mononuclear cell proliferation);GO:0038145(macrophage colony-stimulating factor signaling pathway);GO:0040018(positive regulation of multicellular organism growth);GO:0042488(positive regulation of odontogenesis of dentin-containing tooth);GO:0045657(positive regulation of monocyte differentiation);GO:0046579(positive regulation of Ras protein signal transduction);GO:0048873(homeostasis of number of cells within a tissue);GO:0060444(branching involved in mammary gland duct morphogenesis);GO:0060611(mammary gland fat development);GO:0060763(mammary duct terminal end bud growth);GO:1902228(positive regulation of macrophage colony-stimulating factor signaling pathway);GO:1904141(positive regulation of microglial cell migration) | 04014(Ras signaling pathway);04015(Rap1 signaling pathway);04010(MAPK signaling pathway);04668(TNF signaling pathway);04151(PI3K-Akt signaling pathway);04060(Cytokine-cytokine receptor interaction);04640(Hematopoietic cell lineage);04380(Osteoclast differentiation);05323(Rheumatoid arthritis) | 0.00 | 1 | 599, | 0 | -101 | -99.40 | 3.50 | -3.81 | -5.59 | -0.55 | 3' UTR | 1 | 109915657 | 109922549 | 6893 | 1 |
| chr1 | 111766908 | 111767088 | 181 | ENSG00000064703 | ENST00000534200 | 4491 | DDX20 | DEAD-box helicase 20 [Source:HGNC Symbol;Acc:HGNC:2743] | GO:0003676(nucleic acid binding);GO:0006397(mRNA processing);GO:0008380(RNA splicing);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0005829(cytosol);GO:0016787(hydrolase activity);GO:0004386(helicase activity);GO:0003677(DNA binding);GO:0005515(protein binding);GO:0016604(nuclear body);GO:0005730(nucleolus);GO:0000387(spliceosomal snRNP assembly);GO:0005654(nucleoplasm);GO:0010501(RNA secondary structure unwinding);GO:0043065(positive regulation of apoptotic process);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0005856(cytoskeleton);GO:0004004(ATP-dependent RNA helicase activity);GO:0051170(nuclear import);GO:0032797(SMN complex);GO:0034719(SMN-Sm protein complex);GO:0097504(Gemini of coiled bodies);GO:0006396(RNA processing);GO:0070491(repressing transcription factor binding);GO:0000244(spliceosomal tri-snRNP complex assembly);GO:0019904(protein domain specific binding);GO:0030674(protein binding, bridging);GO:0042826(histone deacetylase binding);GO:0008285(negative regulation of cell proliferation);GO:0045892(negative regulation of transcription, DNA-templated);GO:0048477(oogenesis);GO:0050810(regulation of steroid biosynthetic process);GO:0017053(transcriptional repressor complex);GO:0090571(RNA polymerase II transcription repressor complex) | 03013(RNA transport) | 0.00 | 1 | 181, | 0 | -5.34 | -4.38 | 2.71 | -1.38 | -2.49 | 1.99 | 3' UTR | 1 | 111762417 | 111765830 | 3414 | 1 |
| chr1 | 112456911 | 112457360 | 450 | ENSG00000143079 | ENST00000607039 | 93 | CTTNBP2NL | CTTNBP2 N-terminal like [Source:HGNC Symbol;Acc:HGNC:25330] | GO:0005737(cytoplasm);GO:0006470(protein dephosphorylation);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0015629(actin cytoskeleton);GO:0051721(protein phosphatase 2A binding);GO:0032410(negative regulation of transporter activity);GO:0034763(negative regulation of transmembrane transport) | NA | 0.00 | 1 | 450, | 0 | -55.80 | -54.30 | 3.12 | -1.43 | -2.56 | 0.46 | Exon (ENST00000271277/ENSG00000143079, exon 6 of 6) | 1 | 112456818 | 112463456 | 6639 | 1 |
| chr1 | 147615861 | 147619524 | 3664 | ENSG00000116128 | ENST00000473292 | 4248 | BCL9 | B-cell CLL/lymphoma 9 [Source:HGNC Symbol;Acc:HGNC:1008] | GO:0005634(nucleus);GO:0003713(transcription coactivator activity);GO:0008013(beta-catenin binding);GO:0014908(myotube differentiation involved in skeletal muscle regeneration);GO:0035019(somatic stem cell population maintenance);GO:0035914(skeletal muscle cell differentiation);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0060070(canonical Wnt signaling pathway);GO:0005737(cytoplasm);GO:0005794(Golgi apparatus);GO:0005801(cis-Golgi network);GO:0005515(protein binding);GO:0016055(Wnt signaling pathway);GO:0005654(nucleoplasm);GO:1904837(beta-catenin-TCF complex assembly) | NA | 0.00 | 2 | 42,709, | 0,2955 | -129 | -127 | 4.68 | -1.36 | -2.46 | -0.38 | Exon (ENST00000234739/ENSG00000116128, exon 7 of 10) | 1 | 147611613 | 147615902 | 4290 | 1 |
| chr1 | 149900232 | 149900798 | 567 | ENSG00000178096 | ENST00000369150 | 606 | BOLA1 | bolA family member 1 [Source:HGNC Symbol;Acc:HGNC:24263] | GO:0005739(mitochondrion);GO:0005515(protein binding) | NA | 0.00 | 1 | 567, | 0 | -277 | -274 | 15 | -1.31 | -2.39 | -0.40 | 3' UTR | 1 | 149899626 | 149900548 | 923 | 1 |
| chr1 | 150471599 | 150472347 | 749 | ENSG00000143374 | ENST00000438568 | -15017 | TARS2 | threonyl-tRNA synthetase 2, mitochondrial (putative) [Source:HGNC Symbol;Acc:HGNC:30740] | GO:0005575(cellular_component);GO:0005737(cytoplasm);GO:0006412(translation);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0005739(mitochondrion);GO:0042803(protein homodimerization activity);GO:0016874(ligase activity);GO:0005759(mitochondrial matrix);GO:0004812(aminoacyl-tRNA ligase activity);GO:0006418(tRNA aminoacylation for protein translation);GO:0006450(regulation of translational fidelity);GO:0016876(ligase activity, forming aminoacyl-tRNA and related compounds);GO:0043039(tRNA aminoacylation);GO:0004829(threonine-tRNA ligase activity);GO:0006435(threonyl-tRNA aminoacylation);GO:0002161(aminoacyl-tRNA editing activity);GO:0070159(mitochondrial threonyl-tRNA aminoacylation) | 00970(Aminoacyl-tRNA biosynthesis) | 0.00 | 1 | 749, | 0 | -121 | -119 | 4.21 | -1.44 | -2.57 | 0.33 | Exon (ENST00000492220/ENSG00000163125, exon 11 of 11) | 1 | 150487364 | 150507609 | 20246 | 1 |
| chr1 | 151286971 | 151287420 | 450 | ENSG00000143373 | ENST00000426871 | -64 | ZNF687 | zinc finger protein 687 [Source:HGNC Symbol;Acc:HGNC:29277] | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0005515(protein binding) | NA | 0.00 | 1 | 450, | 0 | -71 | -69.40 | 3.65 | -1.37 | -2.47 | -0.40 | Exon (ENST00000336715/ENSG00000143373, exon 2 of 9) | 1 | 151287484 | 151291903 | 4420 | 1 |
| chr1 | 151291491 | 151291731 | 241 | ENSG00000143373 | ENST00000436614 | 1040 | ZNF687 | zinc finger protein 687 [Source:HGNC Symbol;Acc:HGNC:29277] | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0005515(protein binding) | NA | 0.00 | 1 | 241, | 0 | -32.80 | -31.40 | 8.65 | -1.85 | -3.12 | -1.87 | 3' UTR | 1 | 151290451 | 151291337 | 887 | 1 |
| chr1 | 151759642 | 151759911 | 270 | ENSG00000143436 | ENST00000495867 | 1009 | MRPL9 | mitochondrial ribosomal protein L9 [Source:HGNC Symbol;Acc:HGNC:14277] | GO:0003723(RNA binding);GO:0006412(translation);GO:0005739(mitochondrion);GO:0030529(intracellular ribonucleoprotein complex);GO:0005622(intracellular);GO:0003735(structural constituent of ribosome);GO:0005840(ribosome);GO:0005515(protein binding);GO:0005743(mitochondrial inner membrane);GO:0070125(mitochondrial translational elongation);GO:0070126(mitochondrial translational termination);GO:0005761(mitochondrial ribosome);GO:0005762(mitochondrial large ribosomal subunit) | 03010(Ribosome) | 0.00 | 1 | 270, | 0 | -48.60 | -47.20 | 2.67 | -1.76 | -2.99 | 0.54 | 3' UTR | 1 | 151759884 | 151760920 | 1037 | 2 |
| chr1 | 153661237 | 153661385 | 149 | ENSG00000143553 | ENST00000478558 | 2510 | SNAPIN | SNAP associated protein [Source:HGNC Symbol;Acc:HGNC:17145] | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0006886(intracellular protein transport);GO:0005794(Golgi apparatus);GO:0005765(lysosomal membrane);GO:1904115(axon cytoplasm);GO:0005829(cytosol);GO:0005515(protein binding);GO:0000139(Golgi membrane);GO:0048471(perinuclear region of cytoplasm);GO:0030054(cell junction);GO:0045202(synapse);GO:0031410(cytoplasmic vesicle);GO:0005764(lysosome);GO:0031083(BLOC-1 complex);GO:0006887(exocytosis);GO:0016032(viral process);GO:0008333(endosome to lysosome transport);GO:0008089(anterograde axonal transport);GO:0032438(melanosome organization);GO:0048490(anterograde synaptic vesicle transport);GO:0030672(synaptic vesicle membrane);GO:0031175(neuron projection development);GO:0048489(synaptic vesicle transport);GO:0008021(synaptic vesicle);GO:0030141(secretory granule);GO:0007269(neurotransmitter secretion);GO:0000149(SNARE binding);GO:0007040(lysosome organization);GO:0007042(lysosomal lumen acidification);GO:0007268(chemical synaptic transmission);GO:0008090(retrograde axonal transport);GO:0010977(negative regulation of neuron projection development);GO:0016079(synaptic vesicle exocytosis);GO:0016188(synaptic vesicle maturation);GO:0031629(synaptic vesicle fusion to presynaptic active zone membrane);GO:0032418(lysosome localization);GO:0034629(cellular protein complex localization);GO:0043393(regulation of protein binding);GO:0051604(protein maturation);GO:0072553(terminal button organization);GO:0097352(autophagosome maturation);GO:1902774(late endosome to lysosome transport);GO:1902824(positive regulation of late endosome to lysosome transport);GO:0099078(BORC complex) | NA | 0.00 | 1 | 149, | 0 | -5.38 | -4.42 | 1.28 | -1.34 | -2.43 | -0.51 | 3' UTR | 1 | 153658727 | 153661564 | 2838 | 1 |
| chr1 | 155017775 | 155018522 | 748 | ENSG00000160685 | ENST00000292176 | 3327 | ZBTB7B | zinc finger and BTB domain containing 7B [Source:HGNC Symbol;Acc:HGNC:18668] | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0005654(nucleoplasm);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0005515(protein binding);GO:0007275(multicellular organism development);GO:0030154(cell differentiation);GO:0006366(transcription from RNA polymerase II promoter);GO:0007398(ectoderm development);GO:0010628(positive regulation of gene expression);GO:0000978(RNA polymerase II core promoter proximal region sequence-specific DNA binding);GO:0001077(transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0042803(protein homodimerization activity);GO:0043370(regulation of CD4-positive, alpha-beta T cell differentiation);GO:0043376(regulation of CD8-positive, alpha-beta T cell differentiation);GO:0045944(positive regulation of transcription from RNA polymerase II promoter) | NA | 0.00 | 1 | 748, | 0 | -49.90 | -48.50 | 8.79 | -2.94 | -4.52 | -0.63 | 3' UTR | 1 | 155014448 | 155016981 | 2534 | 1 |
| chr1 | 155737018 | 155738530 | 1513 | ENSG00000132676 | ENST00000491777 | 6 | DAP3 | death associated protein 3 [Source:HGNC Symbol;Acc:HGNC:2673] | GO:0003723(RNA binding);GO:0000166(nucleotide binding);GO:0005654(nucleoplasm);GO:0005739(mitochondrion);GO:0030529(intracellular ribonucleoprotein complex);GO:0005840(ribosome);GO:0005525(GTP binding);GO:0015935(small ribosomal subunit);GO:0006915(apoptotic process);GO:0005515(protein binding);GO:0005761(mitochondrial ribosome);GO:0003735(structural constituent of ribosome);GO:0005763(mitochondrial small ribosomal subunit);GO:0005743(mitochondrial inner membrane);GO:0070125(mitochondrial translational elongation);GO:0070126(mitochondrial translational termination);GO:0097190(apoptotic signaling pathway) | NA | 0.00 | 2 | 46,374, | 0,1139 | -270 | -268 | 9.95 | -2.17 | -3.54 | 0.76 | 3' UTR | 1 | 155737012 | 155739010 | 1999 | 1 |
| chr1 | 156114674 | 156115004 | 331 | ENSG00000160789 | ENST00000368300 | 0 | LMNA | lamin A/C [Source:HGNC Symbol;Acc:HGNC:6636] | GO:0030334(regulation of cell migration);GO:0005634(nucleus);GO:0016607(nuclear speck);GO:0005198(structural molecule activity);GO:0005515(protein binding);GO:0005882(intermediate filament);GO:0005654(nucleoplasm);GO:0005829(cytosol);GO:0031012(extracellular matrix);GO:0031965(nuclear membrane);GO:0005635(nuclear envelope);GO:0036498(IRE1-mediated unfolded protein response);GO:0034504(protein localization to nucleus);GO:0071456(cellular response to hypoxia);GO:0007077(mitotic nuclear envelope disassembly);GO:0005652(nuclear lamina);GO:0007084(mitotic nuclear envelope reassembly);GO:0008285(negative regulation of cell proliferation);GO:0030951(establishment or maintenance of microtubule cytoskeleton polarity);GO:0090343(positive regulation of cell aging);GO:0006997(nucleus organization);GO:0006998(nuclear envelope organization);GO:0010628(positive regulation of gene expression);GO:0035105(sterol regulatory element binding protein import into nucleus);GO:0055015(ventricular cardiac muscle cell development);GO:0072201(negative regulation of mesenchymal cell proliferation);GO:0090201(negative regulation of release of cytochrome c from mitochondria);GO:1900180(regulation of protein localization to nucleus);GO:2001237(negative regulation of extrinsic apoptotic signaling pathway);GO:0005638(lamin filament) | 04210(Apoptosis);05410(Hypertrophic cardiomyopathy (HCM));05412(Arrhythmogenic right ventricular cardiomyopathy (ARVC));05414(Dilated cardiomyopathy (DCM)) | 0 | 1 | 331, | 0 | -4050 | -Inf | 8.29 | -3.57 | -5.31 | -0.20 | 5' UTR | 1 | 156114707 | 156140089 | 25383 | 1 |
| chr1 | 156115274 | 156123200 | 7927 | ENSG00000160789 | ENST00000502357 | 0 | LMNA | lamin A/C [Source:HGNC Symbol;Acc:HGNC:6636] | GO:0030334(regulation of cell migration);GO:0005634(nucleus);GO:0016607(nuclear speck);GO:0005198(structural molecule activity);GO:0005515(protein binding);GO:0005882(intermediate filament);GO:0005654(nucleoplasm);GO:0005829(cytosol);GO:0031012(extracellular matrix);GO:0031965(nuclear membrane);GO:0005635(nuclear envelope);GO:0036498(IRE1-mediated unfolded protein response);GO:0034504(protein localization to nucleus);GO:0071456(cellular response to hypoxia);GO:0007077(mitotic nuclear envelope disassembly);GO:0005652(nuclear lamina);GO:0007084(mitotic nuclear envelope reassembly);GO:0008285(negative regulation of cell proliferation);GO:0030951(establishment or maintenance of microtubule cytoskeleton polarity);GO:0090343(positive regulation of cell aging);GO:0006997(nucleus organization);GO:0006998(nuclear envelope organization);GO:0010628(positive regulation of gene expression);GO:0035105(sterol regulatory element binding protein import into nucleus);GO:0055015(ventricular cardiac muscle cell development);GO:0072201(negative regulation of mesenchymal cell proliferation);GO:0090201(negative regulation of release of cytochrome c from mitochondria);GO:1900180(regulation of protein localization to nucleus);GO:2001237(negative regulation of extrinsic apoptotic signaling pathway);GO:0005638(lamin filament) | 04210(Apoptosis);05410(Hypertrophic cardiomyopathy (HCM));05412(Arrhythmogenic right ventricular cardiomyopathy (ARVC));05414(Dilated cardiomyopathy (DCM)) | 0.00 | 2 | 1,59, | 0,7868 | -33.30 | -31.90 | 1.45 | -1.48 | -2.63 | -0.44 | Exon (ENST00000368301/ENSG00000160789, exon 4 of 13) | 1 | 156123142 | 156134528 | 11387 | 1 |
| chr1 | 156641963 | 156646092 | 4130 | ENSG00000132692 | ENST00000457777 | 0 | BCAN | brevican [Source:HGNC Symbol;Acc:HGNC:23059] | GO:0016020(membrane);GO:0030246(carbohydrate binding);GO:0007155(cell adhesion);GO:0005576(extracellular region);GO:0005578(proteinaceous extracellular matrix);GO:0031225(anchored component of membrane);GO:0005540(hyaluronic acid binding);GO:0043202(lysosomal lumen);GO:0030198(extracellular matrix organization);GO:0030203(glycosaminoglycan metabolic process);GO:0005796(Golgi lumen);GO:0005201(extracellular matrix structural constituent);GO:0001501(skeletal system development);GO:0007417(central nervous system development);GO:0030206(chondroitin sulfate biosynthetic process);GO:0030207(chondroitin sulfate catabolic process);GO:0030208(dermatan sulfate biosynthetic process);GO:0022617(extracellular matrix disassembly);GO:0021766(hippocampus development) | NA | 0.00 | 2 | 313,46, | 0,4084 | -173 | -171 | 7.08 | -3.27 | -4.94 | -0.78 | 5' UTR | 1 | 156642114 | 156647682 | 5569 | 1 |
| chr1 | 158094725 | 158095174 | 450 | ENSG00000183853 | ENST00000368172 | 8045 | KIRREL1 | kirre like nephrin family adhesion molecule 1 [Source:HGNC Symbol;Acc:HGNC:15734] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0048471(perinuclear region of cytoplasm);GO:0070062(extracellular exosome);GO:0007411(axon guidance);GO:0005911(cell-cell junction);GO:0031295(T cell costimulation);GO:0017022(myosin binding);GO:0001933(negative regulation of protein phosphorylation);GO:0007588(excretion);GO:0016337(single organismal cell-cell adhesion);GO:0030838(positive regulation of actin filament polymerization);GO:0031253(cell projection membrane);GO:0043198(dendritic shaft);GO:0045121(membrane raft) | NA | 0.00 | 1 | 450, | 0 | -285 | -283 | 4 | -2.94 | -4.51 | -0.36 | 3' UTR | 1 | 158086680 | 158100262 | 13583 | 1 |
| chr1 | 159782219 | 159782543 | 325 | ENSG00000158716 | ENST00000368107 | 1216 | DUSP23 | dual specificity phosphatase 23 [Source:HGNC Symbol;Acc:HGNC:21480] | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0005654(nucleoplasm);GO:0016787(hydrolase activity);GO:0004725(protein tyrosine phosphatase activity);GO:0008138(protein tyrosine/serine/threonine phosphatase activity);GO:0016791(phosphatase activity);GO:0006470(protein dephosphorylation);GO:0016311(dephosphorylation);GO:0035335(peptidyl-tyrosine dephosphorylation);GO:0005829(cytosol);GO:0004721(phosphoprotein phosphatase activity);GO:0005515(protein binding);GO:0070062(extracellular exosome);GO:0004722(protein serine/threonine phosphatase activity) | NA | 0 | 1 | 325, | 0 | -500 | -Inf | 34.20 | -1.64 | -2.83 | -0.35 | 3' UTR | 1 | 159781003 | 159782543 | 1541 | 1 |
| chr1 | 162383343 | 162383792 | 450 | ENSG00000239887 | ENST00000458626 | 1613 | C1orf226 | chromosome 1 open reading frame 226 [Source:HGNC Symbol;Acc:HGNC:34351] | NA | NA | 0.00 | 1 | 450, | 0 | -37 | -35.60 | 3.09 | -1.39 | -2.51 | 0.57 | 3' UTR | 1 | 162381730 | 162386818 | 5089 | 1 |
| chr1 | 165905929 | 165908270 | 2342 | ENSG00000283936 | ENST00000636291 | 0 | MIR3658 | microRNA 3658 [Source:HGNC Symbol;Acc:HGNC:38963] | NA | NA | 0.00 | 2 | 41,587, | 0,1755 | -108 | -107 | 3.77 | -1.73 | -2.96 | 0.43 | 3' UTR | 1 | 165907921 | 165907976 | 56 | 1 |
| chr1 | 169106354 | 169106683 | 330 | ENSG00000143153 | ENST00000367815 | 31 | ATP1B1 | ATPase Na+/K+ transporting subunit beta 1 [Source:HGNC Symbol;Acc:HGNC:804] | GO:0006810(transport);GO:0016020(membrane);GO:0006811(ion transport);GO:0006813(potassium ion transport);GO:0006814(sodium ion transport);GO:0005890(sodium:potassium-exchanging ATPase complex);GO:0005622(intracellular);GO:0016021(integral component of membrane);GO:0007155(cell adhesion);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0050900(leukocyte migration);GO:0010248(establishment or maintenance of transmembrane electrochemical gradient);GO:0090662(ATP hydrolysis coupled transmembrane transport);GO:0070062(extracellular exosome);GO:0034220(ion transmembrane transport);GO:0042383(sarcolemma);GO:1903278(positive regulation of sodium ion export from cell);GO:1903779(regulation of cardiac conduction);GO:1903561(extracellular vesicle);GO:0051117(ATPase binding);GO:0050821(protein stabilization);GO:0060048(cardiac muscle contraction);GO:0005524(ATP binding);GO:0010107(potassium ion import);GO:0014704(intercalated disc);GO:0016887(ATPase activity);GO:0023026(MHC class II protein complex binding);GO:0005391(sodium:potassium-exchanging ATPase activity);GO:0030955(potassium ion binding);GO:0031402(sodium ion binding);GO:0006883(cellular sodium ion homeostasis);GO:0030007(cellular potassium ion homeostasis);GO:0036376(sodium ion export from cell);GO:0055119(relaxation of cardiac muscle);GO:0086009(membrane repolarization);GO:0086013(membrane repolarization during cardiac muscle cell action potential);GO:0086064(cell communication by electrical coupling involved in cardiac conduction);GO:1990573(potassium ion import across plasma membrane);GO:0008144(drug binding);GO:0001671(ATPase activator activity);GO:0006874(cellular calcium ion homeostasis);GO:0010468(regulation of gene expression);GO:0010882(regulation of cardiac muscle contraction by calcium ion signaling);GO:0032781(positive regulation of ATPase activity);GO:0044861(protein transport into plasma membrane raft);GO:0046034(ATP metabolic process);GO:0072659(protein localization to plasma membrane);GO:1901018(positive regulation of potassium ion transmembrane transporter activity);GO:1903281(positive regulation of calcium:sodium antiporter activity);GO:1903288(positive regulation of potassium ion import);GO:0008022(protein C-terminus binding);GO:0019901(protein kinase binding);GO:0001666(response to hypoxia);GO:0030001(metal ion transport);GO:1903169(regulation of calcium ion transmembrane transport);GO:0005901(caveola);GO:0016323(basolateral plasma membrane);GO:0016324(apical plasma membrane);GO:0043209(myelin sheath) | 04024(cAMP signaling pathway);04022(cGMP - PKG signaling pathway);04911(Insulin secretion);04918(Thyroid hormone synthesis);04919(Thyroid hormone signaling pathway);04925(Aldosterone synthesis and secretion);04260(Cardiac muscle contraction);04261(Adrenergic signaling in cardiomyocytes);04970(Salivary secretion);04971(Gastric acid secretion);04972(Pancreatic secretion);04976(Bile secretion);04973(Carbohydrate digestion and absorption);04974(Protein digestion and absorption);04978(Mineral absorption);04960(Aldosterone-regulated sodium reabsorption);04961(Endocrine and other factor-regulated calcium reabsorption);04964(Proximal tubule bicarbonate reclamation) | 0.00 | 1 | 330, | 0 | -29.40 | -28.10 | 2.75 | -2.11 | -3.46 | -0.87 | 5' UTR | 1 | 169106323 | 169132019 | 25697 | 1 |
| chr1 | 169131346 | 169131466 | 121 | ENSG00000143153 | ENST00000494797 | 23439 | ATP1B1 | ATPase Na+/K+ transporting subunit beta 1 [Source:HGNC Symbol;Acc:HGNC:804] | GO:0006810(transport);GO:0016020(membrane);GO:0006811(ion transport);GO:0006813(potassium ion transport);GO:0006814(sodium ion transport);GO:0005890(sodium:potassium-exchanging ATPase complex);GO:0005622(intracellular);GO:0016021(integral component of membrane);GO:0007155(cell adhesion);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0050900(leukocyte migration);GO:0010248(establishment or maintenance of transmembrane electrochemical gradient);GO:0090662(ATP hydrolysis coupled transmembrane transport);GO:0070062(extracellular exosome);GO:0034220(ion transmembrane transport);GO:0042383(sarcolemma);GO:1903278(positive regulation of sodium ion export from cell);GO:1903779(regulation of cardiac conduction);GO:1903561(extracellular vesicle);GO:0051117(ATPase binding);GO:0050821(protein stabilization);GO:0060048(cardiac muscle contraction);GO:0005524(ATP binding);GO:0010107(potassium ion import);GO:0014704(intercalated disc);GO:0016887(ATPase activity);GO:0023026(MHC class II protein complex binding);GO:0005391(sodium:potassium-exchanging ATPase activity);GO:0030955(potassium ion binding);GO:0031402(sodium ion binding);GO:0006883(cellular sodium ion homeostasis);GO:0030007(cellular potassium ion homeostasis);GO:0036376(sodium ion export from cell);GO:0055119(relaxation of cardiac muscle);GO:0086009(membrane repolarization);GO:0086013(membrane repolarization during cardiac muscle cell action potential);GO:0086064(cell communication by electrical coupling involved in cardiac conduction);GO:1990573(potassium ion import across plasma membrane);GO:0008144(drug binding);GO:0001671(ATPase activator activity);GO:0006874(cellular calcium ion homeostasis);GO:0010468(regulation of gene expression);GO:0010882(regulation of cardiac muscle contraction by calcium ion signaling);GO:0032781(positive regulation of ATPase activity);GO:0044861(protein transport into plasma membrane raft);GO:0046034(ATP metabolic process);GO:0072659(protein localization to plasma membrane);GO:1901018(positive regulation of potassium ion transmembrane transporter activity);GO:1903281(positive regulation of calcium:sodium antiporter activity);GO:1903288(positive regulation of potassium ion import);GO:0008022(protein C-terminus binding);GO:0019901(protein kinase binding);GO:0001666(response to hypoxia);GO:0030001(metal ion transport);GO:1903169(regulation of calcium ion transmembrane transport);GO:0005901(caveola);GO:0016323(basolateral plasma membrane);GO:0016324(apical plasma membrane);GO:0043209(myelin sheath) | 04024(cAMP signaling pathway);04022(cGMP - PKG signaling pathway);04911(Insulin secretion);04918(Thyroid hormone synthesis);04919(Thyroid hormone signaling pathway);04925(Aldosterone synthesis and secretion);04260(Cardiac muscle contraction);04261(Adrenergic signaling in cardiomyocytes);04970(Salivary secretion);04971(Gastric acid secretion);04972(Pancreatic secretion);04976(Bile secretion);04973(Carbohydrate digestion and absorption);04974(Protein digestion and absorption);04978(Mineral absorption);04960(Aldosterone-regulated sodium reabsorption);04961(Endocrine and other factor-regulated calcium reabsorption);04964(Proximal tubule bicarbonate reclamation) | 0.00 | 1 | 121, | 0 | -9.13 | -8.07 | 1.48 | -1.47 | -2.62 | 0.62 | Exon (ENST00000367816/ENSG00000143153, exon 7 of 7) | 1 | 169107907 | 169127399 | 19493 | 1 |
| chr1 | 171532423 | 171535458 | 3036 | ENSG00000117523 | ENST00000495585 | -6372 | PRRC2C | proline rich coiled-coil 2C [Source:HGNC Symbol;Acc:HGNC:24903] | GO:0003723(RNA binding);GO:0016020(membrane);GO:0008022(protein C-terminus binding);GO:0002244(hematopoietic progenitor cell differentiation) | NA | 0.00 | 2 | 539,31, | 0,3005 | -282 | -280 | 5.25 | -10.10 | -12.70 | 0.77 | 3' UTR | 1 | 171541830 | 171593511 | 51682 | 1 |
| chr1 | 171557638 | 171558027 | 390 | ENSG00000117523 | ENST00000495585 | 15808 | PRRC2C | proline rich coiled-coil 2C [Source:HGNC Symbol;Acc:HGNC:24903] | GO:0003723(RNA binding);GO:0016020(membrane);GO:0008022(protein C-terminus binding);GO:0002244(hematopoietic progenitor cell differentiation) | NA | 0.00 | 1 | 390, | 0 | -211 | -209 | 4.08 | -3.39 | -5.09 | 0.47 | Exon (ENST00000367742/ENSG00000117523, exon 19 of 34) | 1 | 171541830 | 171593511 | 51682 | 1 |
| chr1 | 178916503 | 178916713 | 211 | ENSG00000116191 | ENST00000478871 | 22665 | RALGPS2 | Ral GEF with PH domain and SH3 binding motif 2 [Source:HGNC Symbol;Acc:HGNC:30279] | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0007264(small GTPase mediated signal transduction);GO:0005886(plasma membrane);GO:0043547(positive regulation of GTPase activity);GO:0005085(guanyl-nucleotide exchange factor activity);GO:0032485(regulation of Ral protein signal transduction);GO:0005622(intracellular) | NA | 0.00 | 1 | 211, | 0 | -9.38 | -8.32 | 2.79 | -1.64 | -2.83 | 1.43 | 3' UTR | 1 | 178893838 | 178916375 | 22538 | 1 |
| chr1 | 179917842 | 179918291 | 450 | ENSG00000143337 | ENST00000447964 | 13876 | TOR1AIP1 | torsin 1A interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:29456] | GO:0005634(nucleus);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0031965(nuclear membrane);GO:0005515(protein binding);GO:0005637(nuclear inner membrane);GO:0008092(cytoskeletal protein binding);GO:0034504(protein localization to nucleus);GO:0032781(positive regulation of ATPase activity);GO:0001671(ATPase activator activity);GO:0051117(ATPase binding);GO:0005521(lamin binding);GO:0071763(nuclear membrane organization);GO:0005635(nuclear envelope) | NA | 0.00 | 1 | 450, | 0 | -56.40 | -54.90 | 3.34 | -2.62 | -4.12 | 0.57 | 3' UTR | 1 | 179903966 | 179918171 | 14206 | 1 |
| chr1 | 180196307 | 180196845 | 539 | ENSG00000116260 | ENST00000443059 | 0 | QSOX1 | quiescin sulfhydryl oxidase 1 [Source:HGNC Symbol;Acc:HGNC:9756] | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005794(Golgi apparatus);GO:0043231(intracellular membrane-bounded organelle);GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0045454(cell redox homeostasis);GO:0045171(intercellular bridge);GO:0005576(extracellular region);GO:0000139(Golgi membrane);GO:0005615(extracellular space);GO:0003756(protein disulfide isomerase activity);GO:0016972(thiol oxidase activity);GO:0070062(extracellular exosome);GO:0044267(cellular protein metabolic process);GO:0043687(post-translational protein modification);GO:0043312(neutrophil degranulation);GO:0030173(integral component of Golgi membrane);GO:0002576(platelet degranulation);GO:0005788(endoplasmic reticulum lumen);GO:0031093(platelet alpha granule lumen);GO:0016971(flavin-linked sulfhydryl oxidase activity);GO:0016242(negative regulation of macroautophagy);GO:0035580(specific granule lumen);GO:1904724(tertiary granule lumen);GO:0005623(cell) | NA | 0 | 1 | 539, | 0 | -1690 | -Inf | 6.74 | -2.49 | -3.95 | -0.22 | Exon (ENST00000367602/ENSG00000116260, exon 12 of 12) | 1 | 180196682 | 180198016 | 1335 | 1 |
| chr1 | 200848977 | 200849246 | 270 | ENSG00000118200 | ENST00000475326 | -6848 | CAMSAP2 | calmodulin regulated spectrin associated protein family member 2 [Source:HGNC Symbol;Acc:HGNC:29188] | GO:0005737(cytoplasm);GO:0005829(cytosol);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0031175(neuron projection development);GO:0008017(microtubule binding);GO:0005874(microtubule);GO:0005516(calmodulin binding);GO:0005813(centrosome);GO:1990752(microtubule end);GO:0000226(microtubule cytoskeleton organization);GO:0030507(spectrin binding);GO:0051011(microtubule minus-end binding);GO:0033043(regulation of organelle organization) | NA | 0.00 | 1 | 270, | 0 | -12 | -10.90 | 2.22 | -1.42 | -2.54 | 0.92 | Exon (ENST00000358823/ENSG00000118200, exon 11 of 17) | 1 | 200856094 | 200857724 | 1631 | 1 |
| chr1 | 201782690 | 201783465 | 776 | ENSG00000134369 | ENST00000469130 | 60 | NAV1 | neuron navigator 1 [Source:HGNC Symbol;Acc:HGNC:15989] | GO:0005737(cytoplasm);GO:0005856(cytoskeleton);GO:0007275(multicellular organism development);GO:0007399(nervous system development);GO:0030154(cell differentiation);GO:0005874(microtubule);GO:0001578(microtubule bundle formation);GO:0001764(neuron migration);GO:0015630(microtubule cytoskeleton);GO:0043194(axon initial segment) | NA | 0.00 | 2 | 180,60, | 0,716 | -38.60 | -37.20 | 2.46 | -2.44 | -3.89 | -0.78 | Exon (ENST00000367302/ENSG00000134369, exon 8 of 30) | 1 | 201782630 | 201798608 | 15979 | 1 |
| chr1 | 201876152 | 201876421 | 270 | ENSG00000198700 | ENST00000456707 | 1837 | IPO9 | importin 9 [Source:HGNC Symbol;Acc:HGNC:19425] | GO:0006810(transport);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0006886(intracellular protein transport);GO:0015031(protein transport);GO:0008536(Ran GTPase binding);GO:0005515(protein binding);GO:0005829(cytosol);GO:0005635(nuclear envelope);GO:0042393(histone binding);GO:0008565(protein transporter activity);GO:0006606(protein import into nucleus) | NA | 0 | 1 | 270, | 0 | -324 | -Inf | 3.66 | -2.19 | -3.57 | 0.35 | 3' UTR | 1 | 201874315 | 201876373 | 2059 | 1 |
| chr1 | 203165520 | 203165968 | 449 | ENSG00000133055 | ENST00000621380 | 9815 | MYBPH | myosin binding protein H [Source:HGNC Symbol;Acc:HGNC:7552] | GO:0007155(cell adhesion);GO:0005515(protein binding);GO:0051015(actin filament binding);GO:0051371(muscle alpha-actinin binding);GO:0097493(structural molecule activity conferring elasticity);GO:0007015(actin filament organization);GO:0045214(sarcomere organization);GO:0071688(striated muscle myosin thick filament assembly);GO:0005859(muscle myosin complex);GO:0030018(Z disc);GO:0031430(M band);GO:0032982(myosin filament);GO:0008307(structural constituent of muscle);GO:0006942(regulation of striated muscle contraction) | NA | 0 | 1 | 449, | 0 | -357 | -Inf | 4.93 | -3.16 | -4.80 | 0.39 | 3' UTR | 1 | 203167811 | 203175783 | 7973 | 2 |
| chr1 | 206684974 | 206729008 | 44035 | ENSG00000162889 | ENST00000367103 | 0 | MAPKAPK2 | mitogen-activated protein kinase-activated protein kinase 2 [Source:HGNC Symbol;Acc:HGNC:6887] | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0000166(nucleotide binding);GO:0004672(protein kinase activity);GO:0004674(protein serine/threonine kinase activity);GO:0005524(ATP binding);GO:0006468(protein phosphorylation);GO:0005654(nucleoplasm);GO:0016740(transferase activity);GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0005813(centrosome);GO:0005515(protein binding);GO:0006974(cellular response to DNA damage stimulus);GO:0006954(inflammatory response);GO:0005829(cytosol);GO:0070062(extracellular exosome);GO:0043488(regulation of mRNA stability);GO:0032496(response to lipopolysaccharide);GO:0005516(calmodulin binding);GO:0004871(signal transducer activity);GO:0048010(vascular endothelial growth factor receptor signaling pathway);GO:0031572(G2 DNA damage checkpoint);GO:0035924(cellular response to vascular endothelial growth factor stimulus);GO:0006950(response to stress);GO:0018105(peptidyl-serine phosphorylation);GO:1900034(regulation of cellular response to heat);GO:0000187(activation of MAPK activity);GO:0034097(response to cytokine);GO:0002224(toll-like receptor signaling pathway);GO:0007265(Ras protein signal transduction);GO:0032680(regulation of tumor necrosis factor production);GO:0070935(3'-UTR-mediated mRNA stabilization);GO:0006691(leukotriene metabolic process);GO:0000165(MAPK cascade);GO:0004683(calmodulin-dependent protein kinase activity);GO:0009931(calcium-dependent protein serine/threonine kinase activity);GO:0051019(mitogen-activated protein kinase binding);GO:0032675(regulation of interleukin-6 production);GO:0044351(macropinocytosis);GO:0046777(protein autophosphorylation);GO:0038066(p38MAPK cascade);GO:0042535(positive regulation of tumor necrosis factor biosynthetic process);GO:0048255(mRNA stabilization);GO:0048839(inner ear development) | 04010(MAPK signaling pathway);04370(VEGF signaling pathway);04218(Cellular senescence);04722(Neurotrophin signaling pathway);05203(Viral carcinogenesis);05167(Kaposi's sarcoma-associated herpesvirus infection) | 0.00 | 3 | 535,140,75, | 0,43736,43960 | -99.10 | -97.40 | 8.76 | -1.38 | -2.49 | -0.32 | 5' UTR | 1 | 206685037 | 206734283 | 49247 | 1 |
| chr1 | 212444344 | 212446379 | 2036 | ENSG00000117691 | ENST00000472389 | 11353 | NENF | neudesin neurotrophic factor [Source:HGNC Symbol;Acc:HGNC:30384] | GO:0046872(metal ion binding);GO:0005576(extracellular region);GO:0005615(extracellular space);GO:0012505(endomembrane system);GO:0016020(membrane);GO:0032099(negative regulation of appetite);GO:0008083(growth factor activity);GO:0043410(positive regulation of MAPK cascade) | NA | 0.00 | 2 | 99,550, | 0,1486 | -251 | -249 | 14.20 | -1.35 | -2.45 | -0.39 | 3' UTR | 1 | 212432991 | 212435033 | 2043 | 1 |
| chr1 | 214663689 | 214663869 | 181 | ENSG00000117724 | ENST00000469862 | 8541 | CENPF | centromere protein F [Source:HGNC Symbol;Acc:HGNC:1857] | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0015031(protein transport);GO:0005654(nucleoplasm);GO:0051301(cell division);GO:0005694(chromosome);GO:0042803(protein homodimerization activity);GO:0008134(transcription factor binding);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0007049(cell cycle);GO:0007275(multicellular organism development);GO:0048471(perinuclear region of cytoplasm);GO:0030154(cell differentiation);GO:0000922(spindle pole);GO:0005819(spindle);GO:0030496(midbody);GO:0007059(chromosome segregation);GO:0000775(chromosome, centromeric region);GO:0000776(kinetochore);GO:0000777(condensed chromosome kinetochore);GO:0005829(cytosol);GO:0003682(chromatin binding);GO:0008283(cell proliferation);GO:0070840(dynein complex binding);GO:0007517(muscle organ development);GO:0008022(protein C-terminus binding);GO:0071897(DNA biosynthetic process);GO:0007062(sister chromatid cohesion);GO:0045892(negative regulation of transcription, DNA-templated);GO:0000785(chromatin);GO:0016363(nuclear matrix);GO:0005635(nuclear envelope);GO:0000278(mitotic cell cycle);GO:0051310(metaphase plate congression);GO:0007094(mitotic spindle assembly checkpoint);GO:0000940(condensed chromosome outer kinetochore);GO:0042493(response to drug);GO:0051726(regulation of cell cycle);GO:0016202(regulation of striated muscle tissue development);GO:0001822(kidney development);GO:0010389(regulation of G2/M transition of mitotic cell cycle);GO:0021591(ventricular system development);GO:0051382(kinetochore assembly);GO:0005813(centrosome);GO:0005930(axoneme);GO:0036064(ciliary basal body);GO:0045120(pronucleus);GO:0097539(ciliary transition fiber) | NA | 0.00 | 1 | 181, | 0 | -6.50 | -5.51 | 1.97 | -3.14 | -4.78 | 1.82 | 3' UTR | 1 | 214655148 | 214657410 | 2263 | 1 |
| chr1 | 215620254 | 215620493 | 240 | ENSG00000136636 | ENST00000465650 | 1549 | KCTD3 | potassium channel tetramerization domain containing 3 [Source:HGNC Symbol;Acc:HGNC:21305] | GO:0051260(protein homooligomerization) | NA | 0.00 | 1 | 240, | 0 | -21.70 | -20.50 | 2.17 | -2.45 | -3.90 | 0.78 | Exon (ENST00000259154/ENSG00000136636, exon 18 of 18) | 1 | 215618705 | 215620203 | 1499 | 1 |
| chr1 | 223752923 | 223757375 | 4453 | ENSG00000162909 | ENST00000474026 | -249 | CAPN2 | calpain 2 [Source:HGNC Symbol;Acc:HGNC:1479] | GO:0016787(hydrolase activity);GO:0005622(intracellular);GO:0008233(peptidase activity);GO:0008234(cysteine-type peptidase activity);GO:0006508(proteolysis);GO:0004198(calcium-dependent cysteine-type endopeptidase activity);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0046872(metal ion binding);GO:0005829(cytosol);GO:0005509(calcium ion binding);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0005925(focal adhesion);GO:0030425(dendrite);GO:0051493(regulation of cytoskeleton organization);GO:0071230(cellular response to amino acid stimulus);GO:0070062(extracellular exosome);GO:0005783(endoplasmic reticulum);GO:0022617(extracellular matrix disassembly);GO:0045121(membrane raft);GO:0005794(Golgi apparatus);GO:0008092(cytoskeletal protein binding);GO:0051603(proteolysis involved in cellular protein catabolic process);GO:0046982(protein heterodimerization activity);GO:0001666(response to hypoxia);GO:0001824(blastocyst development);GO:0007520(myoblast fusion);GO:0016540(protein autoprocessing);GO:0000785(chromatin);GO:0005634(nucleus);GO:0005764(lysosome);GO:0030864(cortical actin cytoskeleton);GO:0031143(pseudopodium);GO:0097038(perinuclear endoplasmic reticulum) | 04141(Protein processing in endoplasmic reticulum);04210(Apoptosis);04217(Necroptosis);04218(Cellular senescence);04510(Focal adhesion);05010(Alzheimer's disease) | 0.00 | 3 | 34,170,7, | 0,2557,4446 | -73.40 | -71.80 | 1.59 | -1.85 | -3.12 | 0.26 | Exon (ENST00000433674/ENSG00000162909, exon 9 of 21) | 1 | 223757624 | 223776018 | 18395 | 1 |
| chr1 | 231000180 | 231000536 | 357 | ENSG00000173409 | ENST00000459891 | 4420 | ARV1 | ARV1 homolog, fatty acid homeostasis modulator [Source:HGNC Symbol;Acc:HGNC:29561] | GO:0006810(transport);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0006629(lipid metabolic process);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0008202(steroid metabolic process);GO:0006869(lipid transport);GO:0008206(bile acid metabolic process);GO:0030301(cholesterol transport);GO:0090181(regulation of cholesterol metabolic process);GO:0008203(cholesterol metabolic process);GO:0005794(Golgi apparatus);GO:0006695(cholesterol biosynthetic process);GO:0015248(sterol transporter activity);GO:0032383(regulation of intracellular cholesterol transport);GO:0006665(sphingolipid metabolic process);GO:0097036(regulation of plasma membrane sterol distribution);GO:0032541(cortical endoplasmic reticulum) | NA | 0.00 | 1 | 357, | 0 | -70.40 | -68.80 | 9.31 | -1.35 | -2.45 | -0.59 | 3' UTR | 1 | 230995760 | 231000595 | 4836 | 1 |
| chr1 | 234607008 | 234609483 | 2476 | ENSG00000168264 | ENST00000491430 | 0 | IRF2BP2 | interferon regulatory factor 2 binding protein 2 [Source:HGNC Symbol;Acc:HGNC:21729] | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0005654(nucleoplasm);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated) | NA | 0 | 2 | 630,63, | 0,2413 | -415 | -Inf | 3.07 | -1.36 | -2.46 | 0.16 | 3' UTR | 1 | 234607231 | 234608094 | 864 | 2 |
| chr1 | 244053839 | 244054435 | 597 | ENSG00000179456 | ENST00000358704 | 2556 | ZBTB18 | zinc finger and BTB domain containing 18 [Source:HGNC Symbol;Acc:HGNC:13030] | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0005654(nucleoplasm);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0045171(intercellular bridge);GO:0015630(microtubule cytoskeleton);GO:0005515(protein binding);GO:0007275(multicellular organism development);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0043565(sequence-specific DNA binding);GO:0045892(negative regulation of transcription, DNA-templated);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0007519(skeletal muscle tissue development);GO:0000228(nuclear chromosome) | NA | 0.00 | 1 | 597, | 0 | -76 | -74.40 | 2.38 | -4.73 | -6.67 | 0.47 | Exon (ENST00000622512/ENSG00000179456, exon 2 of 2) | 1 | 244051283 | 244057476 | 6194 | 1 |
| chr1 | 244054555 | 244055389 | 835 | ENSG00000179456 | ENST00000358704 | 3272 | ZBTB18 | zinc finger and BTB domain containing 18 [Source:HGNC Symbol;Acc:HGNC:13030] | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0005654(nucleoplasm);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0045171(intercellular bridge);GO:0015630(microtubule cytoskeleton);GO:0005515(protein binding);GO:0007275(multicellular organism development);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0043565(sequence-specific DNA binding);GO:0045892(negative regulation of transcription, DNA-templated);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0007519(skeletal muscle tissue development);GO:0000228(nuclear chromosome) | NA | 0 | 1 | 835, | 0 | -344 | -Inf | 5.83 | -2.44 | -3.89 | 0.27 | 3' UTR | 1 | 244051283 | 244057476 | 6194 | 1 |
| chr1 | 248847393 | 248847961 | 569 | ENSG00000171163 | ENST00000462037 | 6562 | ZNF692 | zinc finger protein 692 [Source:HGNC Symbol;Acc:HGNC:26049] | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0005730(nucleolus);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0000978(RNA polymerase II core promoter proximal region sequence-specific DNA binding);GO:0001078(transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding);GO:0000122(negative regulation of transcription from RNA polymerase II promoter) | NA | 0.00 | 1 | 569, | 0 | -24.10 | -22.90 | 1.76 | -3.20 | -4.86 | -0.52 | Exon (ENST00000306562/ENSG00000171161, exon 4 of 4) | 1 | 248850377 | 248854523 | 4147 | 2 |
| chr1 | 975204 | 975412 | 209 | ENSG00000187642 | ENST00000479361 | 1229 | PERM1 | PPARGC1 and ESRR induced regulator, muscle 1 [Source:HGNC Symbol;Acc:HGNC:28208] | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0014850(response to muscle activity) | NA | 0.00 | 1 | 209, | 0 | -8.76 | -7.71 | 7.95 | -1.49 | -2.64 | -2.53 | 3' UTR | 1 | 975205 | 976641 | 1437 | 2 |
| chr1 | 1335365 | 1336229 | 865 | ENSG00000107404 | ENST00000632445 | 3193 | DVL1 | dishevelled segment polarity protein 1 [Source:HGNC Symbol;Acc:HGNC:3084] | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0035556(intracellular signal transduction);GO:0005829(cytosol);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0007275(multicellular organism development);GO:0016055(Wnt signaling pathway);GO:0031410(cytoplasmic vesicle);GO:0098793(presynapse);GO:0042802(identical protein binding);GO:0006366(transcription from RNA polymerase II promoter);GO:0090090(negative regulation of canonical Wnt signaling pathway);GO:0019899(enzyme binding);GO:0045893(positive regulation of transcription, DNA-templated);GO:0050821(protein stabilization);GO:0060070(canonical Wnt signaling pathway);GO:0099054(presynapse assembly);GO:1905386(positive regulation of protein localization to presynapse);GO:0016328(lateral plasma membrane);GO:0005109(frizzled binding);GO:0019901(protein kinase binding);GO:0001505(regulation of neurotransmitter levels);GO:0006469(negative regulation of protein kinase activity);GO:0007269(neurotransmitter secretion);GO:0007528(neuromuscular junction development);GO:0021915(neural tube development);GO:0030177(positive regulation of Wnt signaling pathway);GO:0032091(negative regulation of protein binding);GO:0032436(positive regulation of proteasomal ubiquitin-dependent protein catabolic process);GO:0034504(protein localization to nucleus);GO:0043113(receptor clustering);GO:0048813(dendrite morphogenesis);GO:0050808(synapse organization);GO:0060071(Wnt signaling pathway, planar cell polarity pathway);GO:0090179(planar cell polarity pathway involved in neural tube closure);GO:1903827(regulation of cellular protein localization);GO:1904886(beta-catenin destruction complex disassembly);GO:0030426(growth cone);GO:0043005(neuron projection);GO:0045202(synapse);GO:0008013(beta-catenin binding);GO:0048365(Rac GTPase binding);GO:0001932(regulation of protein phosphorylation);GO:0001933(negative regulation of protein phosphorylation);GO:0001934(positive regulation of protein phosphorylation);GO:0007409(axonogenesis);GO:0007411(axon guidance);GO:0010976(positive regulation of neuron projection development);GO:0022007(convergent extension involved in neural plate elongation);GO:0031122(cytoplasmic microtubule organization);GO:0035176(social behavior);GO:0035372(protein localization to microtubule);GO:0048668(collateral sprouting);GO:0048675(axon extension);GO:0060029(convergent extension involved in organogenesis);GO:0060134(prepulse inhibition);GO:0060997(dendritic spine morphogenesis);GO:0071340(skeletal muscle acetylcholine-gated channel clustering);GO:0090103(cochlea morphogenesis);GO:0090263(positive regulation of canonical Wnt signaling pathway);GO:2000463(positive regulation of excitatory postsynaptic potential);GO:0005874(microtubule);GO:0014069(postsynaptic density);GO:0015630(microtubule cytoskeleton);GO:0030136(clathrin-coated vesicle);GO:0030424(axon);GO:0030425(dendrite);GO:0043025(neuronal cell body);GO:0043197(dendritic spine);GO:1990909(Wnt signalosome);GO:0005622(intracellular) | 04310(Wnt signaling pathway);04330(Notch signaling pathway);04390(Hippo signaling pathway);04150(mTOR signaling pathway);04550(Signaling pathways regulating pluripotency of stem cells);04916(Melanogenesis);05200(Pathways in cancer);05225(Hepatocellular carcinoma);05226(Gastric cancer);05217(Basal cell carcinoma);05224(Breast cancer);05166(HTLV-I infection);05165(Human papillomavirus infection) | 0.00 | 1 | 865, | 0 | -96.20 | -94.60 | 8.25 | -1.85 | -3.13 | -0.33 | 3' UTR | 1 | 1336412 | 1339422 | 3011 | 2 |
| chr1 | 1598905 | 1599173 | 269 | ENSG00000228594 | ENST00000422725 | 923 | FNDC10 | fibronectin type III domain containing 10 [Source:HGNC Symbol;Acc:HGNC:42951] | GO:0016020(membrane);GO:0016021(integral component of membrane) | NA | 0.00 | 1 | 269, | 0 | -12.10 | -11 | 2.46 | -1.64 | -2.84 | -1.08 | 3' UTR | 1 | 1598012 | 1600096 | 2085 | 2 |
Definition of table:
| Term | Description |
|---|---|
| seqnames | The name of the chromosome (e.g. chr3, chrY, chr2 random) or scaffold (e.g. scaffold10671) |
| start | The starting position of the methylation site in the chromosome or scaffold. |
| end | The ending position of the RNA methylation site in the chromosome or scaffold. |
| width | Peak region of methylation |
| geneId | Defines the ID of gene on which the RNA methylation site locates |
| transcriptId | Defines the ID of transcript on which the RNA methylation site locates |
| distanceToTSS | Distrance from Peak site to transcription start site |
| geneName | Defines the name of gene (gene symbol) on which the RNA methylation site locates |
| GO | Gene ontology term and description http://www.geneontology.org/ |
| KEGG | KEGG pathway annotation and description http://www.kegg.jp/kegg/kegg3a.html |
| score | p-value of the peak |
| blockCount | The number of blocks (exons) the RNA methylation site spans. |
| blockSizes | A comma-separated list of the block sizes. The number of items in this list should correspond to blockCount. |
| blockStarts | A comma-separated list of block starts. All of the blockStart positions should be calculated relative to chromStart. The number of items in this list should correspond to blockCount. |
| lg.p | log10(p-value) of the peak, indicating the significance of the peak as an RNA methylation site. |
| lg.fdr | log10(fdr) of the peak, indicating the significance of the peak as an RNA methylation site after multiple hypothesis correction. |
| fold_enrchment | fold enrichment within the peak in the IP sample compared with the input sample. |
| diff.lg.fdr | results from differential methylation analysis, log10(fdr) of the peak as a differential methylation site between the two experimental conditions tested. |
| diff.lg.p | results from differential methylation analysis, log10(pvalue) of the peak as a differential methylation site between the two experimental conditions tested. |
| diff.log2.fc | results from differential methylation analysis, log2(odds ratio) of the peak as a differential methylation site between the two experimental conditions tested. |
| annotation | Location of peak, such as 3' UTR, first exon... |
| geneStart | The starting position of gene in the chromosome or scaffold. |
| geneEnd | The ending position of gene in the chromosome or scaffold. |
| geneLength | length of gene |
| geneStrand | 1=sense strand or "+"; 2=antisense strand or "-" |
document location:summary/4_diff_peak/*VS*/*VS*_diffPeak.xlsx
peak distribution within different gene contexts:
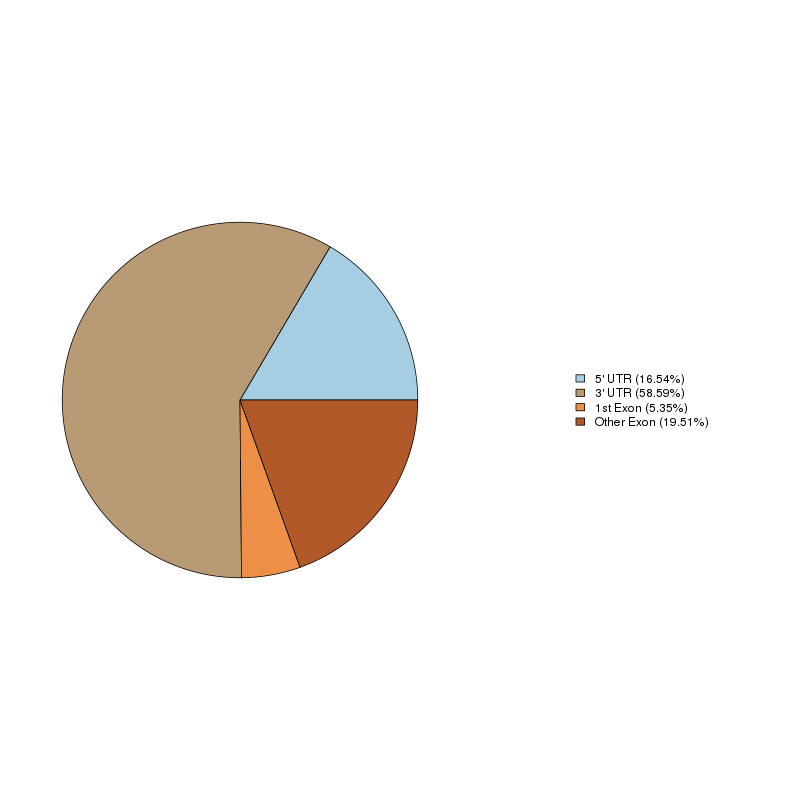
document location:summary/4_diff_peak/*VS*/*VS*_diffPeak_pie.png
Besides differential methylation sites, R package-exomePeak also reports all the detected RNA methylation sites in BED format, based on which the motifs of RNA methylation sites can be detected. The sequence motifs, conjectured to have a biological significance, can be identified in both ChIP-Seq and m6A-seq. The main computational difference is whether to search the reverse complement strand. While a DNA motif may appear on either strand of the two, RNA motif should appear only on the strand where the transcript is located, and thus the strand information should be kept at all times. When the strand information of the RNA fragment is lost in MeRIP-Seq unstranded library construction, it may still be derived from the information of transcripts to which the fragments (reads) are mapped. We use MEME (http://meme-suite.org/) and HOMER (http://homer.ucsd.edu/homer/motif/) to discover RRACH motif of m6A and other noval motifs in methylated sequences.
RRACH motif identification:
de novo motif prediction
document location: summary/5_motif/
StringTie was utilized to assemble all trascripts from mapped reads (bam format) by mapping software Bowtie. Also both gene level and transcript level were estimated by StringTie using FPKM (FPKM=[total_exon_fragments/mapped_reads(millions)×exon_length(kB)]).
| gene_id | gene_name | transcript_id | GO | KEGG | KO_ENTRY | EC | Description | FPKM.M14_OE | FPKM.M14_NC |
| ENSG00000000003 | TSPAN6 | ENST00000612152;ENST00000373020;ENST00000614008;ENST00000496771;ENST00000494424 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0004871(signal transducer activity);GO:0005887(integral component of plasma membrane);GO:0005515(protein binding);GO:0070062(extracellular exosome);GO:0007166(cell surface receptor signaling pathway);GO:0043123(positive regulation of I-kappaB kinase/NF-kappaB signaling);GO:0039532(negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway);GO:1901223(negative regulation of NIK/NF-kappaB signaling);GO:1901223(negative regulation of NIK/NF-kappaB signaling) | NA | NA | NA | tetraspanin 6 [Source:HGNC Symbol;Acc:HGNC:11858] | 4.62 | 4.58 |
| ENSG00000000419 | DPM1 | ENST00000371582 | GO:0005634(nucleus);GO:0016020(membrane);GO:0016740(transferase activity);GO:0016757(transferase activity, transferring glycosyl groups);GO:0005783(endoplasmic reticulum);GO:0005515(protein binding);GO:0006486(protein glycosylation);GO:0006506(GPI anchor biosynthetic process);GO:0005789(endoplasmic reticulum membrane);GO:0004169(dolichyl-phosphate-mannose-protein mannosyltransferase activity);GO:0004582(dolichyl-phosphate beta-D-mannosyltransferase activity);GO:0018279(protein N-linked glycosylation via asparagine);GO:0019348(dolichol metabolic process);GO:0035268(protein mannosylation);GO:0035269(protein O-linked mannosylation);GO:0033185(dolichol-phosphate-mannose synthase complex);GO:0097502(mannosylation);GO:0005537(mannose binding);GO:0043178(alcohol binding);GO:0019673(GDP-mannose metabolic process);GO:0043231(intracellular membrane-bounded organelle) | 00510(N-Glycan biosynthesis) | K00721 | EC:2.4.1.83 | dolichyl-phosphate mannosyltransferase subunit 1, catalytic [Source:HGNC Symbol;Acc:HGNC:3005] | 15.1213920770505 | 19.435594011599 |
| ENSG00000000457 | SCYL3 | ENST00000367771;ENST00000367772;ENST00000423670 | GO:0005737(cytoplasm);GO:0005524(ATP binding);GO:0006468(protein phosphorylation);GO:0005794(Golgi apparatus);GO:0042995(cell projection);GO:0005515(protein binding);GO:0030027(lamellipodium);GO:0016477(cell migration);GO:0004672(protein kinase activity);GO:0004672(protein kinase activity) | NA | NA | NA | SCY1 like pseudokinase 3 [Source:HGNC Symbol;Acc:HGNC:19285] | 1.29 | 1.38 |
| ENSG00000000460 | C1orf112 | ENST00000472795;ENST00000496973;ENST00000359326;ENST00000286031 | NA | NA | NA | NA | chromosome 1 open reading frame 112 [Source:HGNC Symbol;Acc:HGNC:25565] | 2.08 | 2.01 |
| ENSG00000000938 | FGR | ENST00000374005 | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0000166(nucleotide binding);GO:0004672(protein kinase activity);GO:0005524(ATP binding);GO:0006468(protein phosphorylation);GO:0016740(transferase activity);GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0005743(mitochondrial inner membrane);GO:0004713(protein tyrosine kinase activity);GO:0005886(plasma membrane);GO:0005829(cytosol);GO:0005739(mitochondrion);GO:0005856(cytoskeleton);GO:0042995(cell projection);GO:0005515(protein binding);GO:0016235(aggresome);GO:0004715(non-membrane spanning protein tyrosine kinase activity);GO:0007229(integrin-mediated signaling pathway);GO:0002376(immune system process);GO:0045087(innate immune response);GO:0038096(Fc-gamma receptor signaling pathway involved in phagocytosis);GO:0005576(extracellular region);GO:0005758(mitochondrial intermembrane space);GO:0070062(extracellular exosome);GO:0019901(protein kinase binding);GO:0032587(ruffle membrane);GO:0030335(positive regulation of cell migration);GO:0030154(cell differentiation);GO:0050830(defense response to Gram-positive bacterium);GO:0043312(neutrophil degranulation);GO:0008360(regulation of cell shape);GO:0042127(regulation of cell proliferation);GO:0034774(secretory granule lumen);GO:0046777(protein autophosphorylation);GO:0016477(cell migration);GO:0001784(phosphotyrosine residue binding);GO:0009615(response to virus);GO:0045859(regulation of protein kinase activity);GO:0015629(actin cytoskeleton);GO:0014068(positive regulation of phosphatidylinositol 3-kinase signaling);GO:0050764(regulation of phagocytosis);GO:0038083(peptidyl-tyrosine autophosphorylation);GO:0031234(extrinsic component of cytoplasmic side of plasma membrane);GO:0007169(transmembrane receptor protein tyrosine kinase signaling pathway);GO:0018108(peptidyl-tyrosine phosphorylation);GO:0043552(positive regulation of phosphatidylinositol 3-kinase activity);GO:0050715(positive regulation of cytokine secretion);GO:0034987(immunoglobulin receptor binding);GO:0002768(immune response-regulating cell surface receptor signaling pathway);GO:0034988(Fc-gamma receptor I complex binding);GO:0043306(positive regulation of mast cell degranulation);GO:0045088(regulation of innate immune response) | 04062(Chemokine signaling pathway);05169(Epstein-Barr virus infection) | K08891 | EC:2.7.10.2 | FGR proto-oncogene, Src family tyrosine kinase [Source:HGNC Symbol;Acc:HGNC:3697] | 0.09 | 0.09 |
| ENSG00000000971 | CFH | ENST00000367429;ENST00000630130;ENST00000496761;ENST00000466229 | GO:0005515(protein binding);GO:0005576(extracellular region);GO:0002376(immune system process);GO:0045087(innate immune response);GO:0030449(regulation of complement activation);GO:0070062(extracellular exosome);GO:0072562(blood microparticle);GO:0005615(extracellular space);GO:0008201(heparin binding);GO:0006956(complement activation);GO:0006957(complement activation, alternative pathway);GO:0043395(heparan sulfate proteoglycan binding);GO:0043395(heparan sulfate proteoglycan binding) | 04610(Complement and coagulation cascades);05150(Staphylococcus aureus infection);05150(Staphylococcus aureus infection) | K04004 | NA | complement factor H [Source:HGNC Symbol;Acc:HGNC:4883] | 1.48 | 1.13 |
| ENSG00000001036 | FUCA2 | ENST00000002165;ENST00000451668 | GO:0016787(hydrolase activity);GO:0005975(carbohydrate metabolic process);GO:0005515(protein binding);GO:0008152(metabolic process);GO:0005576(extracellular region);GO:0016798(hydrolase activity, acting on glycosyl bonds);GO:0005615(extracellular space);GO:0070062(extracellular exosome);GO:0044267(cellular protein metabolic process);GO:0006004(fucose metabolic process);GO:0043687(post-translational protein modification);GO:0043312(neutrophil degranulation);GO:0005788(endoplasmic reticulum lumen);GO:0035578(azurophil granule lumen);GO:0009617(response to bacterium);GO:0004560(alpha-L-fucosidase activity);GO:0016139(glycoside catabolic process);GO:2000535(regulation of entry of bacterium into host cell);GO:2000535(regulation of entry of bacterium into host cell) | 00511(Other glycan degradation);00511(Other glycan degradation) | K01206 | EC:3.2.1.51 | fucosidase, alpha-L- 2, plasma [Source:HGNC Symbol;Acc:HGNC:4008] | 32.4063505137845 | 32.2261575510204 |
| ENSG00000001084 | GCLC | ENST00000509541;ENST00000229416;ENST00000515580;ENST00000514373;ENST00000514004;ENST00000505294 | GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0016874(ligase activity);GO:0000287(magnesium ion binding);GO:0050880(regulation of blood vessel size);GO:0004357(glutamate-cysteine ligase activity);GO:0006750(glutathione biosynthetic process);GO:0005829(cytosol);GO:0043066(negative regulation of apoptotic process);GO:0006979(response to oxidative stress);GO:0045892(negative regulation of transcription, DNA-templated);GO:0045454(cell redox homeostasis);GO:0009408(response to heat);GO:0006536(glutamate metabolic process);GO:0009725(response to hormone);GO:0050662(coenzyme binding);GO:0043531(ADP binding);GO:0016595(glutamate binding);GO:0046982(protein heterodimerization activity);GO:0006534(cysteine metabolic process);GO:0006749(glutathione metabolic process);GO:0007568(aging);GO:0007584(response to nutrient);GO:0008637(apoptotic mitochondrial changes);GO:0009410(response to xenobiotic stimulus);GO:0014823(response to activity);GO:0019852(L-ascorbic acid metabolic process);GO:0031397(negative regulation of protein ubiquitination);GO:0032436(positive regulation of proteasomal ubiquitin-dependent protein catabolic process);GO:0032869(cellular response to insulin stimulus);GO:0035729(cellular response to hepatocyte growth factor stimulus);GO:0043524(negative regulation of neuron apoptotic process);GO:0044344(cellular response to fibroblast growth factor stimulus);GO:0044752(response to human chorionic gonadotropin);GO:0046685(response to arsenic-containing substance);GO:0046686(response to cadmium ion);GO:0051409(response to nitrosative stress);GO:0051900(regulation of mitochondrial depolarization);GO:0070555(response to interleukin-1);GO:0071260(cellular response to mechanical stimulus);GO:0071333(cellular response to glucose stimulus);GO:0071372(cellular response to follicle-stimulating hormone stimulus);GO:0097069(cellular response to thyroxine stimulus);GO:2000490(negative regulation of hepatic stellate cell activation);GO:2001237(negative regulation of extrinsic apoptotic signaling pathway);GO:0017109(glutamate-cysteine ligase complex);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0016021(integral component of membrane) | 00270(Cysteine and methionine metabolism);00480(Glutathione metabolism);04216(Ferroptosis);04216(Ferroptosis) | K11204 | EC:6.3.2.2 | glutamate-cysteine ligase catalytic subunit [Source:HGNC Symbol;Acc:HGNC:4311] | 5.64 | 5.90 |
| ENSG00000001167 | NFYA | ENST00000341376 | GO:0005634(nucleus);GO:0005654(nucleoplasm);GO:0006355(regulation of transcription, DNA-templated);GO:0006351(transcription, DNA-templated);GO:0003677(DNA binding);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0005515(protein binding);GO:0048511(rhythmic process);GO:0045893(positive regulation of transcription, DNA-templated);GO:0016602(CCAAT-binding factor complex);GO:0006366(transcription from RNA polymerase II promoter);GO:0001046(core promoter sequence-specific DNA binding);GO:0090575(RNA polymerase II transcription factor complex);GO:0045540(regulation of cholesterol biosynthetic process);GO:0044212(transcription regulatory region DNA binding);GO:0000980(RNA polymerase II distal enhancer sequence-specific DNA binding);GO:0032993(protein-DNA complex) | 04612(Antigen processing and presentation);05152(Tuberculosis) | K08064 | NA | nuclear transcription factor Y subunit alpha [Source:HGNC Symbol;Acc:HGNC:7804] | 9.98 | 10.213219 |
| ENSG00000001460 | STPG1 | ENST00000374409;ENST00000468303;ENST00000497384;ENST00000435187;ENST00000498488;ENST00000483528;ENST00000475760 | GO:0005739(mitochondrion);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0006915(apoptotic process);GO:0003674(molecular_function);GO:0005622(intracellular);GO:0043065(positive regulation of apoptotic process);GO:1902110(positive regulation of mitochondrial membrane permeability involved in apoptotic process);GO:0090073(positive regulation of protein homodimerization activity);GO:0090073(positive regulation of protein homodimerization activity) | NA | NA | NA | sperm tail PG-rich repeat containing 1 [Source:HGNC Symbol;Acc:HGNC:28070] | 0.85 | 0.89 |
| ENSG00000001461 | NIPAL3 | ENST00000374399;ENST00000003912;ENST00000488155;ENST00000358028;ENST00000475663 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005515(protein binding);GO:0015095(magnesium ion transmembrane transporter activity);GO:0015693(magnesium ion transport);GO:1903830(magnesium ion transmembrane transport);GO:1903830(magnesium ion transmembrane transport) | NA | NA | NA | NIPA like domain containing 3 [Source:HGNC Symbol;Acc:HGNC:25233] | 0.83 | 0.87 |
| ENSG00000001497 | LAS1L | ENST00000374807;ENST00000374811;ENST00000484069 | GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0005654(nucleoplasm);GO:0005730(nucleolus);GO:0006364(rRNA processing);GO:0030687(preribosome, large subunit precursor);GO:0071339(MLL1 complex);GO:0000470(maturation of LSU-rRNA);GO:0000460(maturation of 5.8S rRNA);GO:0006325(chromatin organization);GO:0000478(endonucleolytic cleavage involved in rRNA processing);GO:0000478(endonucleolytic cleavage involved in rRNA processing) | NA | NA | NA | LAS1 like, ribosome biogenesis factor [Source:HGNC Symbol;Acc:HGNC:25726] | 12.9818208781535 | 15.0097013342279 |
| ENSG00000001561 | ENPP4 | ENST00000321037 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0046872(metal ion binding);GO:0016787(hydrolase activity);GO:0003824(catalytic activity);GO:0008152(metabolic process);GO:0005886(plasma membrane);GO:0070062(extracellular exosome);GO:0043312(neutrophil degranulation);GO:0007596(blood coagulation);GO:0007599(hemostasis);GO:0101003(ficolin-1-rich granule membrane);GO:0047710(bis(5'-adenosyl)-triphosphatase activity);GO:0030194(positive regulation of blood coagulation);GO:0046130(purine ribonucleoside catabolic process) | 00230(Purine metabolism) | K18424 | EC:3.6.1.29 | ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative) [Source:HGNC Symbol;Acc:HGNC:3359] | 1.75 | 2.34 |
| ENSG00000001617 | SEMA3F | ENST00000450338;ENST00000002829 | GO:0005576(extracellular region);GO:0005615(extracellular space);GO:0021675(nerve development);GO:0048846(axon extension involved in axon guidance);GO:0061549(sympathetic ganglion development);GO:0097490(sympathetic neuron projection extension);GO:0097491(sympathetic neuron projection guidance);GO:1902285(semaphorin-plexin signaling pathway involved in neuron projection guidance);GO:0038191(neuropilin binding);GO:0007411(axon guidance);GO:0045499(chemorepellent activity);GO:0001755(neural crest cell migration);GO:0021612(facial nerve structural organization);GO:0021637(trigeminal nerve structural organization);GO:0021785(branchiomotor neuron axon guidance);GO:0036486(ventral trunk neural crest cell migration);GO:0048843(negative regulation of axon extension involved in axon guidance);GO:0050919(negative chemotaxis);GO:1901166(neural crest cell migration involved in autonomic nervous system development);GO:1902287(semaphorin-plexin signaling pathway involved in axon guidance);GO:1902287(semaphorin-plexin signaling pathway involved in axon guidance) | 04360(Axon guidance);04360(Axon guidance) | K06840 | NA | semaphorin 3F [Source:HGNC Symbol;Acc:HGNC:10728] | 0.16 | 0.26 |
| ENSG00000001626 | CFTR | ENST00000003084;ENST00000429014;ENST00000608965 | GO:0016020(membrane);GO:0006811(ion transport);GO:0005254(chloride channel activity);GO:1902476(chloride transmembrane transport);GO:0006821(chloride transport);GO:0006810(transport);GO:0005737(cytoplasm);GO:0016021(integral component of membrane);GO:0055085(transmembrane transport);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0016787(hydrolase activity);GO:0005829(cytosol);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0042626(ATPase activity, coupled to transmembrane movement of substances);GO:0016887(ATPase activity);GO:0005260(intracellular ATPase-gated chloride channel activity);GO:0015106(bicarbonate transmembrane transporter activity);GO:0015108(chloride transmembrane transporter activity);GO:0006695(cholesterol biosynthetic process);GO:0006904(vesicle docking involved in exocytosis);GO:0015701(bicarbonate transport);GO:0030301(cholesterol transport);GO:0035774(positive regulation of insulin secretion involved in cellular response to glucose stimulus);GO:0045921(positive regulation of exocytosis);GO:0048240(sperm capacitation);GO:0051454(intracellular pH elevation);GO:0060081(membrane hyperpolarization);GO:0071320(cellular response to cAMP);GO:1902161(positive regulation of cyclic nucleotide-gated ion channel activity);GO:0005887(integral component of plasma membrane);GO:0016324(apical plasma membrane);GO:0034707(chloride channel complex);GO:0005768(endosome);GO:0070062(extracellular exosome);GO:0061024(membrane organization);GO:0010008(endosome membrane);GO:0016579(protein deubiquitination);GO:0043234(protein complex);GO:0005769(early endosome);GO:0031901(early endosome membrane);GO:0055037(recycling endosome);GO:0055038(recycling endosome membrane);GO:0019899(enzyme binding);GO:0009986(cell surface);GO:0030165(PDZ domain binding);GO:0005765(lysosomal membrane);GO:0019869(chloride channel inhibitor activity);GO:0043225(ATPase-coupled anion transmembrane transporter activity);GO:0099133(ATP hydrolysis coupled anion transmembrane transport);GO:1904322(cellular response to forskolin);GO:0030665(clathrin-coated vesicle membrane);GO:0030660(Golgi-associated vesicle membrane);GO:0031205(endoplasmic reticulum Sec complex);GO:0017081(chloride channel regulator activity);GO:0035377(transepithelial water transport);GO:0050891(multicellular organismal water homeostasis);GO:1902943(positive regulation of voltage-gated chloride channel activity);GO:1902943(positive regulation of voltage-gated chloride channel activity) | 02010(ABC transporters);04024(cAMP signaling pathway);04152(AMPK signaling pathway);04530(Tight junction);04971(Gastric acid secretion);04972(Pancreatic secretion);04976(Bile secretion);05110(Vibrio cholerae infection);05110(Vibrio cholerae infection) | K05031 | EC:3.6.3.49 | cystic fibrosis transmembrane conductance regulator [Source:HGNC Symbol;Acc:HGNC:1884] | 0.02 | 0.03 |
| ENSG00000001629 | ANKIB1 | ENST00000265742;ENST00000413588 | GO:0046872(metal ion binding);GO:0016740(transferase activity);GO:0004842(ubiquitin-protein transferase activity);GO:0005515(protein binding);GO:0016567(protein ubiquitination);GO:0005737(cytoplasm);GO:0000151(ubiquitin ligase complex);GO:0061630(ubiquitin protein ligase activity);GO:0042787(protein ubiquitination involved in ubiquitin-dependent protein catabolic process);GO:0000209(protein polyubiquitination);GO:0032436(positive regulation of proteasomal ubiquitin-dependent protein catabolic process);GO:0031624(ubiquitin conjugating enzyme binding);GO:0031624(ubiquitin conjugating enzyme binding) | NA | NA | NA | ankyrin repeat and IBR domain containing 1 [Source:HGNC Symbol;Acc:HGNC:22215] | 4.90 | 5.67 |
| ENSG00000001630 | CYP51A1 | ENST00000003100 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0046872(metal ion binding);GO:0005783(endoplasmic reticulum);GO:0004497(monooxygenase activity);GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0005506(iron ion binding);GO:0016705(oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen);GO:0020037(heme binding);GO:0008398(sterol 14-demethylase activity);GO:0033488(cholesterol biosynthetic process via 24,25-dihydrolanosterol);GO:0070988(demethylation);GO:0043231(intracellular membrane-bounded organelle);GO:0006629(lipid metabolic process);GO:0005789(endoplasmic reticulum membrane);GO:0006694(steroid biosynthetic process);GO:0008202(steroid metabolic process);GO:0016126(sterol biosynthetic process);GO:0008203(cholesterol metabolic process);GO:0005886(plasma membrane);GO:0006695(cholesterol biosynthetic process);GO:0031090(organelle membrane);GO:0045540(regulation of cholesterol biosynthetic process);GO:0016125(sterol metabolic process) | 00100(Steroid biosynthesis) | K05917 | EC:1.14.13.70 | cytochrome P450 family 51 subfamily A member 1 [Source:HGNC Symbol;Acc:HGNC:2649] | 18.4869383946981 | 18.9701182474227 |
| ENSG00000001631 | KRIT1 | ENST00000394507;ENST00000470309 | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0005886(plasma membrane);GO:0050790(regulation of catalytic activity);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0005615(extracellular space);GO:0030054(cell junction);GO:0005874(microtubule);GO:0008017(microtubule binding);GO:2000114(regulation of establishment of cell polarity);GO:0005546(phosphatidylinositol-4,5-bisphosphate binding);GO:0030695(GTPase regulator activity);GO:0005911(cell-cell junction);GO:0032403(protein complex binding);GO:0001525(angiogenesis);GO:0001937(negative regulation of endothelial cell proliferation);GO:0007264(small GTPase mediated signal transduction);GO:0010596(negative regulation of endothelial cell migration);GO:0016525(negative regulation of angiogenesis);GO:0045454(cell redox homeostasis);GO:2000352(negative regulation of endothelial cell apoptotic process);GO:0032092(positive regulation of protein binding);GO:0043234(protein complex);GO:0043234(protein complex) | 04015(Rap1 signaling pathway);04015(Rap1 signaling pathway) | K17705 | NA | KRIT1, ankyrin repeat containing [Source:HGNC Symbol;Acc:HGNC:1573] | 1.91 | 1.96 |
| ENSG00000002016 | RAD52 | ENST00000358495;ENST00000468231;ENST00000536177 | GO:0005634(nucleus);GO:0003677(DNA binding);GO:0006281(DNA repair);GO:0005515(protein binding);GO:0006310(DNA recombination);GO:0006974(cellular response to DNA damage stimulus);GO:0000724(double-strand break repair via homologous recombination);GO:0042802(identical protein binding);GO:0005654(nucleoplasm);GO:0043234(protein complex);GO:0000730(DNA recombinase assembly);GO:0045002(double-strand break repair via single-strand annealing);GO:0051260(protein homooligomerization);GO:0006302(double-strand break repair);GO:0032993(protein-DNA complex);GO:0034599(cellular response to oxidative stress);GO:0003697(single-stranded DNA binding);GO:0010792(DNA double-strand break processing involved in repair via single-strand annealing);GO:2000819(regulation of nucleotide-excision repair);GO:2000819(regulation of nucleotide-excision repair) | 03440(Homologous recombination);03440(Homologous recombination) | K10873 | NA | RAD52 homolog, DNA repair protein [Source:HGNC Symbol;Acc:HGNC:9824] | 0.73 | 0.77 |
| ENSG00000002079 | MYH16 | ENST00000439784;ENST00000452010 | NA | NA | NA | NA | myosin heavy chain 16 pseudogene [Source:HGNC Symbol;Acc:HGNC:31038] | 1.93 | 2.30 |
| ENSG00000002330 | BAD | ENST00000394532;ENST00000493798;ENST00000492141;ENST00000544271 | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0005739(mitochondrion);GO:0005829(cytosol);GO:0006915(apoptotic process);GO:0005515(protein binding);GO:0005741(mitochondrial outer membrane);GO:0008656(cysteine-type endopeptidase activator activity involved in apoptotic process);GO:0019903(protein phosphatase binding);GO:0030346(protein phosphatase 2B binding);GO:0043422(protein kinase B binding);GO:0046982(protein heterodimerization activity);GO:0071889(14-3-3 protein binding);GO:0001666(response to hypoxia);GO:0001836(release of cytochrome c from mitochondria);GO:0006007(glucose catabolic process);GO:0006919(activation of cysteine-type endopeptidase activity involved in apoptotic process);GO:0007283(spermatogenesis);GO:0008283(cell proliferation);GO:0008625(extrinsic apoptotic signaling pathway via death domain receptors);GO:0008630(intrinsic apoptotic signaling pathway in response to DNA damage);GO:0009725(response to hormone);GO:0009749(response to glucose);GO:0010033(response to organic substance);GO:0010918(positive regulation of mitochondrial membrane potential);GO:0014070(response to organic cyclic compound);GO:0019050(suppression by virus of host apoptotic process);GO:0019221(cytokine-mediated signaling pathway);GO:0021987(cerebral cortex development);GO:0032024(positive regulation of insulin secretion);GO:0032355(response to estradiol);GO:0032570(response to progesterone);GO:0033133(positive regulation of glucokinase activity);GO:0033574(response to testosterone);GO:0034201(response to oleic acid);GO:0035774(positive regulation of insulin secretion involved in cellular response to glucose stimulus);GO:0042493(response to drug);GO:0042542(response to hydrogen peroxide);GO:0042593(glucose homeostasis);GO:0042981(regulation of apoptotic process);GO:0043065(positive regulation of apoptotic process);GO:0043200(response to amino acid);GO:0043281(regulation of cysteine-type endopeptidase activity involved in apoptotic process);GO:0044342(type B pancreatic cell proliferation);GO:0045471(response to ethanol);GO:0045579(positive regulation of B cell differentiation);GO:0045582(positive regulation of T cell differentiation);GO:0046031(ADP metabolic process);GO:0046034(ATP metabolic process);GO:0051384(response to glucocorticoid);GO:0051592(response to calcium ion);GO:0060139(positive regulation of apoptotic process by virus);GO:0060154(cellular process regulating host cell cycle in response to virus);GO:0071247(cellular response to chromate);GO:0071396(cellular response to lipid);GO:0097190(apoptotic signaling pathway);GO:0097192(extrinsic apoptotic signaling pathway in absence of ligand);GO:1901216(positive regulation of neuron death);GO:1902220(positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress);GO:2000078(positive regulation of type B pancreatic cell development);GO:0008289(lipid binding);GO:0019901(protein kinase binding);GO:0010508(positive regulation of autophagy);GO:0071260(cellular response to mechanical stimulus);GO:0097191(extrinsic apoptotic signaling pathway);GO:0090200(positive regulation of release of cytochrome c from mitochondria);GO:0005543(phospholipid binding);GO:0043280(positive regulation of cysteine-type endopeptidase activity involved in apoptotic process);GO:0097193(intrinsic apoptotic signaling pathway);GO:0071456(cellular response to hypoxia);GO:1900740(positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway);GO:0050679(positive regulation of epithelial cell proliferation);GO:0001844(protein insertion into mitochondrial membrane involved in apoptotic signaling pathway);GO:2001244(positive regulation of intrinsic apoptotic signaling pathway);GO:0071316(cellular response to nicotine);GO:0046902(regulation of mitochondrial membrane permeability);GO:0046931(pore complex assembly);GO:0045862(positive regulation of proteolysis);GO:0097202(activation of cysteine-type endopeptidase activity);GO:0045918(negative regulation of cytolysis);GO:0045918(negative regulation of cytolysis) | 04014(Ras signaling pathway);04012(ErbB signaling pathway);04370(VEGF signaling pathway);04024(cAMP signaling pathway);04022(cGMP - PKG signaling pathway);04151(PI3K-Akt signaling pathway);04140(Autophagy - animal);04210(Apoptosis);04510(Focal adhesion);04910(Insulin signaling pathway);04919(Thyroid hormone signaling pathway);04722(Neurotrophin signaling pathway);05200(Pathways in cancer);05203(Viral carcinogenesis);05210(Colorectal cancer);05212(Pancreatic cancer);05225(Hepatocellular carcinoma);05221(Acute myeloid leukemia);05220(Chronic myeloid leukemia);05218(Melanoma);05211(Renal cell carcinoma);05215(Prostate cancer);05213(Endometrial cancer);05223(Non-small cell lung cancer);05010(Alzheimer's disease);05014(Amyotrophic lateral sclerosis (ALS));05152(Tuberculosis);05161(Hepatitis B);05160(Hepatitis C);05165(Human papillomavirus infection);05145(Toxoplasmosis);01521(EGFR tyrosine kinase inhibitor resistance);01524(Platinum drug resistance);01522(Endocrine resistance);01522(Endocrine resistance) | K02158 | NA | BCL2 associated agonist of cell death [Source:HGNC Symbol;Acc:HGNC:936] | 22.606028868267 | 24.080227941452 |
| ENSG00000002549 | LAP3 | ENST00000226299;ENST00000508497;ENST00000509583;ENST00000503467 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0016787(hydrolase activity);GO:0005622(intracellular);GO:0008233(peptidase activity);GO:0006508(proteolysis);GO:0030145(manganese ion binding);GO:0030496(midbody);GO:0008235(metalloexopeptidase activity);GO:0005925(focal adhesion);GO:0004177(aminopeptidase activity);GO:0070062(extracellular exosome);GO:0019538(protein metabolic process);GO:0005739(mitochondrion);GO:0005802(trans-Golgi network);GO:0005802(trans-Golgi network) | 00330(Arginine and proline metabolism);00480(Glutathione metabolism);00480(Glutathione metabolism) | NA | NA | leucine aminopeptidase 3 [Source:HGNC Symbol;Acc:HGNC:18449] | 1.82 | 1.94 |
| ENSG00000002586 | CD99 | ENST00000381192;ENST00000449611;ENST00000482293 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0007155(cell adhesion);GO:0005925(focal adhesion);GO:0050776(regulation of immune response);GO:0005886(plasma membrane);GO:0005887(integral component of plasma membrane);GO:0005515(protein binding);GO:0005737(cytoplasm);GO:0005737(cytoplasm) | 04514(Cell adhesion molecules (CAMs));04670(Leukocyte transendothelial migration);04670(Leukocyte transendothelial migration) | K06520 | NA | CD99 molecule (Xg blood group) [Source:HGNC Symbol;Acc:HGNC:7082] | 17.1329192335566 | 14.8691970523752 |
| ENSG00000002587 | HS3ST1 | ENST00000002596 | GO:0016740(transferase activity);GO:0005794(Golgi apparatus);GO:0008146(sulfotransferase activity);GO:0016021(integral component of membrane);GO:0005796(Golgi lumen);GO:0006024(glycosaminoglycan biosynthetic process);GO:0008467([heparan sulfate]-glucosamine 3-sulfotransferase 1 activity) | 00534(Glycosaminoglycan biosynthesis - heparan sulfate / heparin) | K01024 | EC:2.8.2.23 | heparan sulfate-glucosamine 3-sulfotransferase 1 [Source:HGNC Symbol;Acc:HGNC:5194] | 1.64 | 1.73 |
| ENSG00000002726 | AOC1 | ENST00000493429 | GO:0046872(metal ion binding);GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0008270(zinc ion binding);GO:0042803(protein homodimerization activity);GO:0005886(plasma membrane);GO:0005576(extracellular region);GO:0005615(extracellular space);GO:0005509(calcium ion binding);GO:0005507(copper ion binding);GO:0005777(peroxisome);GO:0005923(bicellular tight junction);GO:0008201(heparin binding);GO:0070062(extracellular exosome);GO:0004872(receptor activity);GO:0006805(xenobiotic metabolic process);GO:0043312(neutrophil degranulation);GO:0008131(primary amine oxidase activity);GO:0048038(quinone binding);GO:0009308(amine metabolic process);GO:0035580(specific granule lumen);GO:0008144(drug binding);GO:0032403(protein complex binding);GO:0046677(response to antibiotic);GO:0042493(response to drug);GO:0071504(cellular response to heparin);GO:0071280(cellular response to copper ion);GO:0071420(cellular response to histamine);GO:0052597(diamine oxidase activity);GO:0052598(histamine oxidase activity);GO:0052599(methylputrescine oxidase activity);GO:0052600(propane-1,3-diamine oxidase activity);GO:0035874(cellular response to copper ion starvation);GO:0097185(cellular response to azide) | 00330(Arginine and proline metabolism);00340(Histidine metabolism);00380(Tryptophan metabolism) | NA | NA | amine oxidase, copper containing 1 [Source:HGNC Symbol;Acc:HGNC:80] | 0 | 0.05 |
| ENSG00000002745 | WNT16 | ENST00000222462 | GO:0005102(receptor binding);GO:0005576(extracellular region);GO:0007275(multicellular organism development);GO:0016055(Wnt signaling pathway);GO:0005578(proteinaceous extracellular matrix);GO:0005615(extracellular space);GO:0010628(positive regulation of gene expression);GO:0005737(cytoplasm);GO:0014068(positive regulation of phosphatidylinositol 3-kinase signaling);GO:0030182(neuron differentiation);GO:0046330(positive regulation of JNK cascade);GO:0005109(frizzled binding);GO:0045165(cell fate commitment);GO:0090399(replicative senescence);GO:0060548(negative regulation of cell death);GO:0090403(oxidative stress-induced premature senescence);GO:0030216(keratinocyte differentiation);GO:0003408(optic cup formation involved in camera-type eye development);GO:0043616(keratinocyte proliferation);GO:0046849(bone remodeling);GO:0060317(cardiac epithelial to mesenchymal transition) | 04310(Wnt signaling pathway);04390(Hippo signaling pathway);04150(mTOR signaling pathway);04550(Signaling pathways regulating pluripotency of stem cells);04916(Melanogenesis);05200(Pathways in cancer);05202(Transcriptional misregulation in cancers);05205(Proteoglycans in cancer);05225(Hepatocellular carcinoma);05226(Gastric cancer);05217(Basal cell carcinoma);05224(Breast cancer);05166(HTLV-I infection);05165(Human papillomavirus infection) | K01558 | NA | Wnt family member 16 [Source:HGNC Symbol;Acc:HGNC:16267] | 0.01 | 0.00 |
| ENSG00000002746 | HECW1 | ENST00000395891;ENST00000471043 | GO:0005737(cytoplasm);GO:0005829(cytosol);GO:0016740(transferase activity);GO:0016567(protein ubiquitination);GO:0004842(ubiquitin-protein transferase activity);GO:0061630(ubiquitin protein ligase activity);GO:0043161(proteasome-mediated ubiquitin-dependent protein catabolic process);GO:0090090(negative regulation of canonical Wnt signaling pathway);GO:0042787(protein ubiquitination involved in ubiquitin-dependent protein catabolic process);GO:0042787(protein ubiquitination involved in ubiquitin-dependent protein catabolic process) | NA | NA | NA | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 [Source:HGNC Symbol;Acc:HGNC:22195] | 0.00 | 0.01 |
| ENSG00000002822 | MAD1L1 | ENST00000481633;ENST00000265854;ENST00000406869;ENST00000437877;ENST00000444373;ENST00000421113;ENST00000491858;ENST00000464742;ENST00000459731;ENST00000486340;ENST00000469871;ENST00000429625 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0005829(cytosol);GO:0051301(cell division);GO:0005694(chromosome);GO:0015629(actin cytoskeleton);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0007049(cell cycle);GO:0005643(nuclear pore);GO:0005813(centrosome);GO:0005815(microtubule organizing center);GO:0005819(spindle);GO:0000775(chromosome, centromeric region);GO:0000776(kinetochore);GO:0000777(condensed chromosome kinetochore);GO:0007094(mitotic spindle assembly checkpoint);GO:0007062(sister chromatid cohesion);GO:0007093(mitotic cell cycle checkpoint);GO:0072686(mitotic spindle);GO:0051315(attachment of mitotic spindle microtubules to kinetochore);GO:0090235(regulation of metaphase plate congression);GO:0042130(negative regulation of T cell proliferation);GO:0048538(thymus development);GO:1901990(regulation of mitotic cell cycle phase transition);GO:0000922(spindle pole);GO:0000922(spindle pole) | 04110(Cell cycle);04914(Progesterone-mediated oocyte maturation);05203(Viral carcinogenesis);05203(Viral carcinogenesis) | K06638 | NA | MAD1 mitotic arrest deficient like 1 [Source:HGNC Symbol;Acc:HGNC:6762] | 4.12 | 3.64 |
| ENSG00000002834 | LASP1 | ENST00000433206;ENST00000318008;ENST00000584106;ENST00000443937;ENST00000419929;ENST00000579123 | GO:0005737(cytoplasm);GO:0006810(transport);GO:0046872(metal ion binding);GO:0034220(ion transmembrane transport);GO:0009967(positive regulation of signal transduction);GO:0003779(actin binding);GO:0006811(ion transport);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0015075(ion transmembrane transporter activity);GO:0051015(actin filament binding);GO:0005925(focal adhesion);GO:0030864(cortical actin cytoskeleton);GO:0045296(cadherin binding);GO:0070062(extracellular exosome);GO:0005938(cell cortex);GO:0005070(SH3/SH2 adaptor activity);GO:0005070(SH3/SH2 adaptor activity) | NA | NA | NA | LIM and SH3 protein 1 [Source:HGNC Symbol;Acc:HGNC:6513] | 192.271292394137 | 191.150864861351 |
| ENSG00000002919 | SNX11 | ENST00000581298;ENST00000393405;ENST00000580875;ENST00000583320 | GO:0005622(intracellular);GO:0006886(intracellular protein transport);GO:0035091(phosphatidylinositol binding);GO:0006810(transport);GO:0016020(membrane);GO:0015031(protein transport);GO:0005515(protein binding);GO:0008289(lipid binding);GO:0005768(endosome);GO:0019898(extrinsic component of membrane);GO:0006897(endocytosis);GO:0016050(vesicle organization);GO:1901981(phosphatidylinositol phosphate binding);GO:1901981(phosphatidylinositol phosphate binding) | NA | NA | NA | sorting nexin 11 [Source:HGNC Symbol;Acc:HGNC:14975] | 6.84 | 7.03 |
| ENSG00000002933 | TMEM176A | ENST00000462826 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:2001199(negative regulation of dendritic cell differentiation) | NA | NA | NA | transmembrane protein 176A [Source:HGNC Symbol;Acc:HGNC:24930] | 0 | 0.00 |
| ENSG00000003056 | M6PR | ENST00000539143;ENST00000541507;ENST00000537621;ENST00000543258;ENST00000540837 | GO:0016021(integral component of membrane);GO:0005537(mannose binding);GO:0015578(mannose transmembrane transporter activity);GO:0015761(mannose transport);GO:0000323(lytic vacuole);GO:0006810(transport);GO:0016020(membrane);GO:0005765(lysosomal membrane);GO:0048471(perinuclear region of cytoplasm);GO:0007165(signal transduction);GO:0005764(lysosome);GO:0005886(plasma membrane);GO:0061024(membrane organization);GO:0005887(integral component of plasma membrane);GO:0005802(trans-Golgi network);GO:0032588(trans-Golgi network membrane);GO:0005515(protein binding);GO:0030133(transport vesicle);GO:0004888(transmembrane signaling receptor activity);GO:0030665(clathrin-coated vesicle membrane);GO:0030904(retromer complex);GO:0006898(receptor-mediated endocytosis);GO:0005768(endosome);GO:0008333(endosome to lysosome transport);GO:1905394(retromer complex binding);GO:0033299(secretion of lysosomal enzymes);GO:0005770(late endosome);GO:0005770(late endosome) | 04145(Phagosome);04142(Lysosome);04142(Lysosome) | NA | NA | mannose-6-phosphate receptor, cation dependent [Source:HGNC Symbol;Acc:HGNC:6752] | 37.8938845541435 | 38.733813394493 |
| ENSG00000003096 | KLHL13 | ENST00000371876;ENST00000540167 | GO:0016567(protein ubiquitination);GO:0051301(cell division);GO:0007049(cell cycle);GO:0005829(cytosol);GO:0043687(post-translational protein modification);GO:0004842(ubiquitin-protein transferase activity);GO:0000910(cytokinesis);GO:0031463(Cul3-RING ubiquitin ligase complex);GO:0030496(midbody);GO:0030496(midbody) | 04120(Ubiquitin mediated proteolysis);04120(Ubiquitin mediated proteolysis) | K10447 | NA | kelch like family member 13 [Source:HGNC Symbol;Acc:HGNC:22931] | 0.59 | 0.84 |
| ENSG00000003137 | CYP26B1 | ENST00000001146 | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0046872(metal ion binding);GO:0004497(monooxygenase activity);GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0005506(iron ion binding);GO:0016705(oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen);GO:0020037(heme binding);GO:0006805(xenobiotic metabolic process);GO:0043231(intracellular membrane-bounded organelle);GO:0060349(bone morphogenesis);GO:0016709(oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen);GO:0031090(organelle membrane);GO:0006766(vitamin metabolic process);GO:0001972(retinoic acid binding);GO:0008401(retinoic acid 4-hydroxylase activity);GO:0016125(sterol metabolic process);GO:0034653(retinoic acid catabolic process);GO:0048387(negative regulation of retinoic acid receptor signaling pathway);GO:0030326(embryonic limb morphogenesis);GO:0007283(spermatogenesis);GO:0001709(cell fate determination);GO:0001768(establishment of T cell polarity);GO:0006954(inflammatory response);GO:0007140(male meiotic nuclear division);GO:0009954(proximal/distal pattern formation);GO:0010628(positive regulation of gene expression);GO:0042573(retinoic acid metabolic process);GO:0043587(tongue morphogenesis);GO:0045580(regulation of T cell differentiation);GO:0048384(retinoic acid receptor signaling pathway);GO:0048385(regulation of retinoic acid receptor signaling pathway);GO:0061436(establishment of skin barrier);GO:0070268(cornification);GO:0071300(cellular response to retinoic acid);GO:2001037(positive regulation of tongue muscle cell differentiation) | 00830(Retinol metabolism) | K12664 | NA | cytochrome P450 family 26 subfamily B member 1 [Source:HGNC Symbol;Acc:HGNC:20581] | 0.47 | 0.42 |
| ENSG00000003147 | ICA1 | ENST00000402384;ENST00000406470;ENST00000455539;ENST00000422063;ENST00000490041;ENST00000446305 | GO:0006810(transport);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0043231(intracellular membrane-bounded organelle);GO:0005829(cytosol);GO:0005794(Golgi apparatus);GO:0019904(protein domain specific binding);GO:0005515(protein binding);GO:0000139(Golgi membrane);GO:0030054(cell junction);GO:0045202(synapse);GO:0031410(cytoplasmic vesicle);GO:0050796(regulation of insulin secretion);GO:0030667(secretory granule membrane);GO:0030672(synaptic vesicle membrane);GO:0003674(molecular_function);GO:0006836(neurotransmitter transport);GO:0030658(transport vesicle membrane);GO:0016021(integral component of membrane);GO:0016021(integral component of membrane) | 04940(Type I diabetes mellitus);04940(Type I diabetes mellitus) | K19863 | NA | islet cell autoantigen 1 [Source:HGNC Symbol;Acc:HGNC:5343] | 0.53 | 0.55 |
| ENSG00000003249 | DBNDD1 | ENST00000002501;ENST00000623401;ENST00000392973;ENST00000568838 | GO:0005737(cytoplasm);GO:0030672(synaptic vesicle membrane);GO:0031175(neuron projection development);GO:0031175(neuron projection development) | NA | NA | NA | dysbindin domain containing 1 [Source:HGNC Symbol;Acc:HGNC:28455] | 11.6134778747652 | 11.6739960402839 |
| ENSG00000003393 | ALS2 | ENST00000264276;ENST00000439495;ENST00000482891;ENST00000467448;ENST00000496244;ENST00000409632;ENST00000410052 | GO:0005829(cytosol);GO:0042803(protein homodimerization activity);GO:0005089(Rho guanyl-nucleotide exchange factor activity);GO:0035023(regulation of Rho protein signal transduction);GO:0005515(protein binding);GO:0005085(guanyl-nucleotide exchange factor activity);GO:0030425(dendrite);GO:0048812(neuron projection morphogenesis);GO:0030027(lamellipodium);GO:0030426(growth cone);GO:0043539(protein serine/threonine kinase activator activity);GO:0043547(positive regulation of GTPase activity);GO:0017112(Rab guanyl-nucleotide exchange factor activity);GO:0061024(membrane organization);GO:0043234(protein complex);GO:0005769(early endosome);GO:0071902(positive regulation of protein serine/threonine kinase activity);GO:0001726(ruffle);GO:0031982(vesicle);GO:0045860(positive regulation of protein kinase activity);GO:0005813(centrosome);GO:0017137(Rab GTPase binding);GO:0007032(endosome organization);GO:0030676(Rac guanyl-nucleotide exchange factor activity);GO:0051036(regulation of endosome size);GO:0001662(behavioral fear response);GO:0001701(in utero embryonic development);GO:0001881(receptor recycling);GO:0006979(response to oxidative stress);GO:0007409(axonogenesis);GO:0007528(neuromuscular junction development);GO:0007626(locomotory behavior);GO:0008104(protein localization);GO:0008219(cell death);GO:0016050(vesicle organization);GO:0016197(endosomal transport);GO:0016601(Rac protein signal transduction);GO:0035022(positive regulation of Rac protein signal transduction);GO:0035249(synaptic transmission, glutamatergic);GO:0043087(regulation of GTPase activity);GO:0005737(cytoplasm);GO:0014069(postsynaptic density);GO:0016020(membrane);GO:0030424(axon);GO:0043005(neuron projection);GO:0043025(neuronal cell body);GO:0043197(dendritic spine);GO:0043231(intracellular membrane-bounded organelle);GO:0043231(intracellular membrane-bounded organelle) | 05014(Amyotrophic lateral sclerosis (ALS));05014(Amyotrophic lateral sclerosis (ALS)) | K04575 | NA | ALS2, alsin Rho guanine nucleotide exchange factor [Source:HGNC Symbol;Acc:HGNC:443] | 5.07 | 5.75 |
| ENSG00000003400 | CASP10 | ENST00000286186;ENST00000272879;ENST00000374650 | GO:0016787(hydrolase activity);GO:0042981(regulation of apoptotic process);GO:0005515(protein binding);GO:0008233(peptidase activity);GO:0008234(cysteine-type peptidase activity);GO:0006508(proteolysis);GO:0006915(apoptotic process);GO:0004197(cysteine-type endopeptidase activity);GO:0005829(cytosol);GO:0031625(ubiquitin protein ligase binding);GO:0043123(positive regulation of I-kappaB kinase/NF-kappaB signaling);GO:0097190(apoptotic signaling pathway);GO:0007166(cell surface receptor signaling pathway);GO:0097200(cysteine-type endopeptidase activity involved in execution phase of apoptosis);GO:0097194(execution phase of apoptosis);GO:0097342(ripoptosome);GO:0031265(CD95 death-inducing signaling complex);GO:0035877(death effector domain binding);GO:0097199(cysteine-type endopeptidase activity involved in apoptotic signaling pathway);GO:0097199(cysteine-type endopeptidase activity involved in apoptotic signaling pathway) | 04668(TNF signaling pathway);04210(Apoptosis);04622(RIG-I-like receptor signaling pathway);05152(Tuberculosis);05161(Hepatitis B);05161(Hepatitis B) | K04400 | EC:3.4.22.63 | caspase 10 [Source:HGNC Symbol;Acc:HGNC:1500] | 1.40 | 1.38 |
| ENSG00000003402 | CFLAR | ENST00000309955;ENST00000342795;ENST00000395148;ENST00000441224;ENST00000423241;ENST00000425030;ENST00000440180;ENST00000457277;ENST00000494258;ENST00000470178;ENST00000470664 | GO:0005737(cytoplasm);GO:0042981(regulation of apoptotic process);GO:0043085(positive regulation of catalytic activity);GO:0005515(protein binding);GO:0008234(cysteine-type peptidase activity);GO:0006508(proteolysis);GO:0006915(apoptotic process);GO:0004197(cysteine-type endopeptidase activity);GO:0002020(protease binding);GO:0005123(death receptor binding);GO:0032403(protein complex binding);GO:0001666(response to hypoxia);GO:0010667(negative regulation of cardiac muscle cell apoptotic process);GO:0010976(positive regulation of neuron projection development);GO:0032869(cellular response to insulin stimulus);GO:0033574(response to testosterone);GO:0043066(negative regulation of apoptotic process);GO:0070374(positive regulation of ERK1 and ERK2 cascade);GO:0071364(cellular response to epidermal growth factor stimulus);GO:0071392(cellular response to estradiol stimulus);GO:0071456(cellular response to hypoxia);GO:0071549(cellular response to dexamethasone stimulus);GO:0071732(cellular response to nitric oxide);GO:0072126(positive regulation of glomerular mesangial cell proliferation);GO:1903055(positive regulation of extracellular matrix organization);GO:1903427(negative regulation of reactive oxygen species biosynthetic process);GO:1903845(negative regulation of cellular response to transforming growth factor beta stimulus);GO:1903944(negative regulation of hepatocyte apoptotic process);GO:1904036(negative regulation of epithelial cell apoptotic process);GO:2000347(positive regulation of hepatocyte proliferation);GO:0031264(death-inducing signaling complex);GO:0031265(CD95 death-inducing signaling complex);GO:0045121(membrane raft);GO:0005829(cytosol);GO:0016032(viral process);GO:0043123(positive regulation of I-kappaB kinase/NF-kappaB signaling);GO:1902041(regulation of extrinsic apoptotic signaling pathway via death domain receptors);GO:1902042(negative regulation of extrinsic apoptotic signaling pathway via death domain receptors);GO:0051092(positive regulation of NF-kappaB transcription factor activity);GO:0008047(enzyme activator activity);GO:0097200(cysteine-type endopeptidase activity involved in execution phase of apoptosis);GO:0007519(skeletal muscle tissue development);GO:0014732(skeletal muscle atrophy);GO:0014842(regulation of skeletal muscle satellite cell proliferation);GO:0014866(skeletal myofibril assembly);GO:0043403(skeletal muscle tissue regeneration);GO:0060544(regulation of necroptotic process);GO:0097194(execution phase of apoptosis);GO:1901740(negative regulation of myoblast fusion);GO:2001237(negative regulation of extrinsic apoptotic signaling pathway);GO:0097342(ripoptosome);GO:0097342(ripoptosome) | 04064(NF-kappa B signaling pathway);04668(TNF signaling pathway);04140(Autophagy - animal);04210(Apoptosis);04217(Necroptosis);05142(Chagas disease (American trypanosomiasis));05142(Chagas disease (American trypanosomiasis)) | K04724 | NA | CASP8 and FADD like apoptosis regulator [Source:HGNC Symbol;Acc:HGNC:1876] | 1.49 | 1.50 |
| ENSG00000003436 | TFPI | ENST00000392365;ENST00000409676;ENST00000421427;ENST00000420747 | GO:0005615(extracellular space);GO:0016020(membrane);GO:0004867(serine-type endopeptidase inhibitor activity);GO:0010951(negative regulation of endopeptidase activity);GO:0005783(endoplasmic reticulum);GO:0005576(extracellular region);GO:0031225(anchored component of membrane);GO:0005886(plasma membrane);GO:0030414(peptidase inhibitor activity);GO:0010466(negative regulation of peptidase activity);GO:0043231(intracellular membrane-bounded organelle);GO:0009986(cell surface);GO:0007596(blood coagulation);GO:0007599(hemostasis);GO:0031090(organelle membrane);GO:0004866(endopeptidase inhibitor activity);GO:0007598(blood coagulation, extrinsic pathway);GO:0032355(response to estradiol);GO:0032496(response to lipopolysaccharide);GO:0071222(cellular response to lipopolysaccharide);GO:0071347(cellular response to interleukin-1);GO:0071347(cellular response to interleukin-1) | 04610(Complement and coagulation cascades);04610(Complement and coagulation cascades) | K03909 | NA | tissue factor pathway inhibitor [Source:HGNC Symbol;Acc:HGNC:11760] | 2.63 | 3.02 |
| ENSG00000003509 | NDUFAF7 | ENST00000455230;ENST00000474154;ENST00000441905 | GO:0008168(methyltransferase activity);GO:0016740(transferase activity);GO:0032259(methylation);GO:0005739(mitochondrion);GO:0005615(extracellular space);GO:0005515(protein binding);GO:0032981(mitochondrial respiratory chain complex I assembly);GO:0005759(mitochondrial matrix);GO:0019899(enzyme binding);GO:0035243(protein-arginine omega-N symmetric methyltransferase activity);GO:0019918(peptidyl-arginine methylation, to symmetrical-dimethyl arginine);GO:0046034(ATP metabolic process);GO:0046034(ATP metabolic process) | NA | NA | NA | NADH:ubiquinone oxidoreductase complex assembly factor 7 [Source:HGNC Symbol;Acc:HGNC:28816] | 1.84 | 2.03 |
| ENSG00000003756 | RBM5 | ENST00000347869;ENST00000464087;ENST00000492472;ENST00000437500;ENST00000496179;ENST00000471995;ENST00000494360;ENST00000474470;ENST00000479275 | GO:0003676(nucleic acid binding);GO:0003723(RNA binding);GO:0006397(mRNA processing);GO:0008380(RNA splicing);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0005654(nucleoplasm);GO:0042981(regulation of apoptotic process);GO:0005515(protein binding);GO:0005681(spliceosomal complex);GO:0006915(apoptotic process);GO:0000381(regulation of alternative mRNA splicing, via spliceosome);GO:0003729(mRNA binding);GO:0008285(negative regulation of cell proliferation);GO:0043065(positive regulation of apoptotic process);GO:0003677(DNA binding);GO:0000398(mRNA splicing, via spliceosome);GO:0006396(RNA processing);GO:0000245(spliceosomal complex assembly);GO:0000245(spliceosomal complex assembly) | NA | NA | NA | RNA binding motif protein 5 [Source:HGNC Symbol;Acc:HGNC:9902] | 2.09 | 2.24 |
| ENSG00000003987 | MTMR7 | ENST00000180173 | GO:0005737(cytoplasm);GO:0016787(hydrolase activity);GO:0004725(protein tyrosine phosphatase activity);GO:0016791(phosphatase activity);GO:0016311(dephosphorylation);GO:0035335(peptidyl-tyrosine dephosphorylation);GO:0005515(protein binding);GO:0046856(phosphatidylinositol dephosphorylation);GO:0005829(cytosol);GO:0052866(phosphatidylinositol phosphate phosphatase activity);GO:0006661(phosphatidylinositol biosynthetic process);GO:0006470(protein dephosphorylation);GO:0004438(phosphatidylinositol-3-phosphatase activity);GO:0052629(phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity);GO:0046855(inositol phosphate dephosphorylation);GO:0016020(membrane) | 00562(Inositol phosphate metabolism);04070(Phosphatidylinositol signaling system) | K18083 | EC:3.1.3.64 3.1.3.95 | myotubularin related protein 7 [Source:HGNC Symbol;Acc:HGNC:7454] | 0.14 | 0.12 |
| ENSG00000003989 | SLC7A2 | ENST00000494857;ENST00000470360 | GO:0006810(transport);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005886(plasma membrane);GO:0030054(cell junction);GO:0015171(amino acid transmembrane transporter activity);GO:0003333(amino acid transmembrane transport);GO:1990822(basic amino acid transmembrane transport);GO:0006865(amino acid transport);GO:1903826(arginine transmembrane transport);GO:0005887(integral component of plasma membrane);GO:0015174(basic amino acid transmembrane transporter activity);GO:0006520(cellular amino acid metabolic process);GO:0000064(L-ornithine transmembrane transporter activity);GO:0005289(high-affinity arginine transmembrane transporter activity);GO:0005292(high-affinity lysine transmembrane transporter activity);GO:0015181(arginine transmembrane transporter activity);GO:0015189(L-lysine transmembrane transporter activity);GO:0061459(L-arginine transmembrane transporter activity);GO:0097626(low-affinity L-arginine transmembrane transporter activity);GO:0097627(high-affinity L-ornithine transmembrane transporter activity);GO:0002537(nitric oxide production involved in inflammatory response);GO:0006809(nitric oxide biosynthetic process);GO:0015809(arginine transport);GO:0042116(macrophage activation);GO:0043030(regulation of macrophage activation);GO:0050727(regulation of inflammatory response);GO:0097638(L-arginine import across plasma membrane);GO:0097639(L-lysine import across plasma membrane);GO:0097640(L-ornithine import across plasma membrane);GO:1902023(L-arginine transport);GO:1903352(L-ornithine transmembrane transport);GO:1903352(L-ornithine transmembrane transport) | NA | NA | NA | solute carrier family 7 member 2 [Source:HGNC Symbol;Acc:HGNC:11060] | 1.72 | 1.86 |
| ENSG00000004059 | ARF5 | ENST00000000233;ENST00000463733;ENST00000467281;ENST00000489673 | GO:0006810(transport);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0015031(protein transport);GO:0005525(GTP binding);GO:0007264(small GTPase mediated signal transduction);GO:0000166(nucleotide binding);GO:0005794(Golgi apparatus);GO:0005622(intracellular);GO:0048471(perinuclear region of cytoplasm);GO:0016192(vesicle-mediated transport);GO:0005515(protein binding);GO:0070062(extracellular exosome);GO:0005886(plasma membrane);GO:0003924(GTPase activity);GO:0006890(retrograde vesicle-mediated transport, Golgi to ER);GO:0006890(retrograde vesicle-mediated transport, Golgi to ER) | 04144(Endocytosis);04144(Endocytosis) | K07940 | NA | ADP ribosylation factor 5 [Source:HGNC Symbol;Acc:HGNC:658] | 32.6310664975949 | 42.6214167316943 |
| ENSG00000004139 | SARM1 | ENST00000379061;ENST00000585482 | GO:0005737(cytoplasm);GO:0007165(signal transduction);GO:0005739(mitochondrion);GO:0042995(cell projection);GO:0005515(protein binding);GO:0030054(cell junction);GO:0045202(synapse);GO:0007399(nervous system development);GO:0030154(cell differentiation);GO:0005874(microtubule);GO:0002376(immune system process);GO:0045087(innate immune response);GO:0030425(dendrite);GO:0030424(axon);GO:0005829(cytosol);GO:0034128(negative regulation of MyD88-independent toll-like receptor signaling pathway);GO:0048814(regulation of dendrite morphogenesis);GO:1901214(regulation of neuron death);GO:0009749(response to glucose);GO:0042981(regulation of apoptotic process);GO:0015630(microtubule cytoskeleton);GO:0031315(extrinsic component of mitochondrial outer membrane);GO:0031315(extrinsic component of mitochondrial outer membrane) | NA | NA | NA | sterile alpha and TIR motif containing 1 [Source:HGNC Symbol;Acc:HGNC:17074] | 0.28 | 0.39 |
| ENSG00000004142 | POLDIP2 | ENST00000540200 | GO:0005634(nucleus);GO:0003677(DNA binding);GO:0005515(protein binding);GO:0042645(mitochondrial nucleoid);GO:0005739(mitochondrion);GO:0016242(negative regulation of macroautophagy);GO:0045931(positive regulation of mitotic cell cycle);GO:0070584(mitochondrion morphogenesis) | NA | NA | NA | DNA polymerase delta interacting protein 2 [Source:HGNC Symbol;Acc:HGNC:23781] | 85.148712 | 81.740311 |
| ENSG00000004399 | PLXND1 | ENST00000324093;ENST00000501038;ENST00000504524;ENST00000506979;ENST00000508630;ENST00000512807;ENST00000503166;ENST00000504767 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0007165(signal transduction);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0007275(multicellular organism development);GO:0017154(semaphorin receptor activity);GO:0071526(semaphorin-plexin signaling pathway);GO:0005887(integral component of plasma membrane);GO:0030027(lamellipodium);GO:0008360(regulation of cell shape);GO:0043087(regulation of GTPase activity);GO:0030334(regulation of cell migration);GO:0019904(protein domain specific binding);GO:0001525(angiogenesis);GO:0007162(negative regulation of cell adhesion);GO:0050772(positive regulation of axonogenesis);GO:0043542(endothelial cell migration);GO:0007416(synapse assembly);GO:0045765(regulation of angiogenesis);GO:0001569(branching involved in blood vessel morphogenesis);GO:0003151(outflow tract morphogenesis);GO:0003279(cardiac septum development);GO:0032092(positive regulation of protein binding);GO:0035904(aorta development);GO:0060666(dichotomous subdivision of terminal units involved in salivary gland branching);GO:0060976(coronary vasculature development);GO:0060976(coronary vasculature development) | NA | NA | NA | plexin D1 [Source:HGNC Symbol;Acc:HGNC:9107] | 9.94 | 8.62 |
| ENSG00000004455 | AK2 | ENST00000629371;ENST00000491241;ENST00000487289 | GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0016740(transferase activity);GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0006139(nucleobase-containing compound metabolic process);GO:0005739(mitochondrion);GO:0019205(nucleobase-containing compound kinase activity);GO:0004017(adenylate kinase activity);GO:0016776(phosphotransferase activity, phosphate group as acceptor);GO:0006172(ADP biosynthetic process);GO:0046939(nucleotide phosphorylation);GO:0005758(mitochondrial intermembrane space);GO:0070062(extracellular exosome);GO:0015949(nucleobase-containing small molecule interconversion);GO:0046033(AMP metabolic process);GO:0046034(ATP metabolic process);GO:0005743(mitochondrial inner membrane);GO:0036126(sperm flagellum);GO:0097226(sperm mitochondrial sheath);GO:0097226(sperm mitochondrial sheath) | 00230(Purine metabolism);00730(Thiamine metabolism);00730(Thiamine metabolism) | NA | NA | adenylate kinase 2 [Source:HGNC Symbol;Acc:HGNC:362] | 27.7112631290379 | 26.2483904843723 |
| ENSG00000004468 | CD38 | ENST00000226279 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0016740(transferase activity);GO:0016787(hydrolase activity);GO:0045893(positive regulation of transcription, DNA-templated);GO:0005886(plasma membrane);GO:0030890(positive regulation of B cell proliferation);GO:0070062(extracellular exosome);GO:0003953(NAD+ nucleosidase activity);GO:0050135(NAD(P)+ nucleosidase activity);GO:0043066(negative regulation of apoptotic process);GO:0007165(signal transduction);GO:0045892(negative regulation of transcription, DNA-templated);GO:0050853(B cell receptor signaling pathway);GO:0097190(apoptotic signaling pathway);GO:0042493(response to drug);GO:0019674(NAD metabolic process);GO:0016849(phosphorus-oxygen lyase activity);GO:0061809(NAD+ nucleotidase, cyclic ADP-ribose generating);GO:0016798(hydrolase activity, acting on glycosyl bonds);GO:0001666(response to hypoxia);GO:0007204(positive regulation of cytosolic calcium ion concentration);GO:0007565(female pregnancy);GO:0009725(response to hormone);GO:0030307(positive regulation of cell growth);GO:0032024(positive regulation of insulin secretion);GO:0032355(response to estradiol);GO:0032526(response to retinoic acid);GO:0032570(response to progesterone);GO:0033194(response to hydroperoxide);GO:0034097(response to cytokine);GO:0045779(negative regulation of bone resorption);GO:0045907(positive regulation of vasoconstriction);GO:0060292(long term synaptic depression);GO:0070555(response to interleukin-1);GO:0005634(nucleus);GO:0009986(cell surface);GO:0043231(intracellular membrane-bounded organelle) | 00760(Nicotinate and nicotinamide metabolism);04020(Calcium signaling pathway);04640(Hematopoietic cell lineage);04921(Oxytocin signaling pathway);04970(Salivary secretion);04972(Pancreatic secretion);05169(Epstein-Barr virus infection) | K01242 | EC:3.2.2.6 2.4.99.20 | CD38 molecule [Source:HGNC Symbol;Acc:HGNC:1667] | 0.00 | 0.01 |
| ENSG00000004478 | FKBP4 | ENST00000001008;ENST00000540260;ENST00000538622;ENST00000543037;ENST00000539181 | GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016853(isomerase activity);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0003755(peptidyl-prolyl cis-trans isomerase activity);GO:0000413(protein peptidyl-prolyl isomerization);GO:0005739(mitochondrion);GO:0005856(cytoskeleton);GO:0042995(cell projection);GO:0005515(protein binding);GO:0005528(FK506 binding);GO:0005874(microtubule);GO:0031072(heat shock protein binding);GO:0030424(axon);GO:0044295(axonal growth cone);GO:0070062(extracellular exosome);GO:0061077(chaperone-mediated protein folding);GO:1900034(regulation of cellular response to heat);GO:0010977(negative regulation of neuron projection development);GO:0006457(protein folding);GO:0030674(protein binding, bridging);GO:0005524(ATP binding);GO:0005525(GTP binding);GO:0032767(copper-dependent protein binding);GO:0035259(glucocorticoid receptor binding);GO:0048156(tau protein binding);GO:0051219(phosphoprotein binding);GO:0006463(steroid hormone receptor complex assembly);GO:0006825(copper ion transport);GO:0007566(embryo implantation);GO:0030521(androgen receptor signaling pathway);GO:0030850(prostate gland development);GO:0031111(negative regulation of microtubule polymerization or depolymerization);GO:0031115(negative regulation of microtubule polymerization);GO:0031503(protein complex localization);GO:0046661(male sex differentiation);GO:0048608(reproductive structure development);GO:0043005(neuron projection);GO:0043025(neuronal cell body);GO:0043234(protein complex);GO:0048471(perinuclear region of cytoplasm);GO:0048471(perinuclear region of cytoplasm) | 04915(Estrogen signaling pathway);04915(Estrogen signaling pathway) | K09571 | EC:5.2.1.8 | FK506 binding protein 4 [Source:HGNC Symbol;Acc:HGNC:3720] | 16.7106222355212 | 16.9041828025097 |
| ENSG00000004487 | KDM1A | ENST00000356634;ENST00000400181;ENST00000494920;ENST00000602503 | GO:0051091(positive regulation of sequence-specific DNA binding transcription factor activity);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0043433(negative regulation of sequence-specific DNA binding transcription factor activity);GO:0005634(nucleus);GO:0005654(nucleoplasm);GO:0016491(oxidoreductase activity);GO:0050660(flavin adenine dinucleotide binding);GO:0055114(oxidation-reduction process);GO:0006355(regulation of transcription, DNA-templated);GO:0006351(transcription, DNA-templated);GO:0016569(covalent chromatin modification);GO:0016575(histone deacetylation);GO:0003677(DNA binding);GO:0005515(protein binding);GO:0007275(multicellular organism development);GO:0030374(ligand-dependent nuclear receptor transcription coactivator activity);GO:0003682(chromatin binding);GO:0006357(regulation of transcription from RNA polymerase II promoter);GO:0000784(nuclear chromosome, telomeric region);GO:0043234(protein complex);GO:0032454(histone demethylase activity (H3-K9 specific));GO:0033169(histone H3-K9 demethylation);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0000790(nuclear chromatin);GO:0045892(negative regulation of transcription, DNA-templated);GO:0004407(histone deacetylase activity);GO:0044212(transcription regulatory region DNA binding);GO:0043426(MRF binding);GO:0032452(histone demethylase activity);GO:0019899(enzyme binding);GO:0032091(negative regulation of protein binding);GO:0007596(blood coagulation);GO:0050681(androgen receptor binding);GO:0043518(negative regulation of DNA damage response, signal transduction by p53 class mediator);GO:0002039(p53 binding);GO:1902166(negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator);GO:0008134(transcription factor binding);GO:0032451(demethylase activity);GO:0032453(histone demethylase activity (H3-K4 specific));GO:0034648(histone demethylase activity (H3-dimethyl-K4 specific));GO:0042162(telomeric DNA binding);GO:0061752(telomeric repeat-containing RNA binding);GO:0006482(protein demethylation);GO:0010569(regulation of double-strand break repair via homologous recombination);GO:0033184(positive regulation of histone ubiquitination);GO:0034644(cellular response to UV);GO:0034720(histone H3-K4 demethylation);GO:0043392(negative regulation of DNA binding);GO:0055001(muscle cell development);GO:0071480(cellular response to gamma radiation);GO:1903827(regulation of cellular protein localization);GO:0005667(transcription factor complex);GO:1990391(DNA repair complex);GO:0000380(alternative mRNA splicing, via spliceosome);GO:0010976(positive regulation of neuron projection development);GO:0014070(response to organic cyclic compound);GO:0021987(cerebral cortex development);GO:0035563(positive regulation of chromatin binding);GO:0042551(neuron maturation);GO:0045793(positive regulation of cell size);GO:0046098(guanine metabolic process);GO:0060992(response to fungicide);GO:0071320(cellular response to cAMP);GO:0071320(cellular response to cAMP) | NA | NA | NA | lysine demethylase 1A [Source:HGNC Symbol;Acc:HGNC:29079] | 11.2141982338546 | 12.557098750138 |
| ENSG00000004534 | RBM6 | ENST00000442092;ENST00000443081;ENST00000454079;ENST00000434592;ENST00000488807;ENST00000446471;ENST00000478026 | GO:0003676(nucleic acid binding);GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005515(protein binding);GO:0003677(DNA binding);GO:0006396(RNA processing);GO:0006396(RNA processing) | NA | NA | NA | RNA binding motif protein 6 [Source:HGNC Symbol;Acc:HGNC:9903] | 3.91 | 3.49 |
| ENSG00000004660 | CAMKK1 | ENST00000381769;ENST00000348335;ENST00000158166;ENST00000573483 | GO:0000166(nucleotide binding);GO:0004672(protein kinase activity);GO:0004674(protein serine/threonine kinase activity);GO:0005524(ATP binding);GO:0006468(protein phosphorylation);GO:0016740(transferase activity);GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0005515(protein binding);GO:0005516(calmodulin binding);GO:0035556(intracellular signal transduction);GO:0005829(cytosol);GO:0018105(peptidyl-serine phosphorylation);GO:0018107(peptidyl-threonine phosphorylation);GO:0045860(positive regulation of protein kinase activity);GO:0004683(calmodulin-dependent protein kinase activity);GO:0004683(calmodulin-dependent protein kinase activity) | 05034(Alcoholism);05034(Alcoholism) | K00908 | EC:2.7.11.17 | calcium/calmodulin dependent protein kinase kinase 1 [Source:HGNC Symbol;Acc:HGNC:1469] | 2.35 | 2.21 |
| ENSG00000004700 | RECQL | ENST00000444129 | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0016020(membrane);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0005654(nucleoplasm);GO:0016787(hydrolase activity);GO:0004386(helicase activity);GO:0008026(ATP-dependent helicase activity);GO:0032508(DNA duplex unwinding);GO:0003677(DNA binding);GO:0005515(protein binding);GO:0006310(DNA recombination);GO:0005737(cytoplasm);GO:0004003(ATP-dependent DNA helicase activity);GO:0006281(DNA repair);GO:0000724(double-strand break repair via homologous recombination);GO:0003678(DNA helicase activity);GO:0009378(four-way junction helicase activity);GO:0043140(ATP-dependent 3'-5' DNA helicase activity);GO:0005694(chromosome);GO:0000733(DNA strand renaturation);GO:0036310(annealing helicase activity) | NA | NA | NA | RecQ like helicase [Source:HGNC Symbol;Acc:HGNC:9948] | 4.45 | 4.65 |
| ENSG00000004766 | VPS50 | ENST00000251739;ENST00000544910;ENST00000471188;ENST00000495039;ENST00000458707 | GO:0006810(transport);GO:0016020(membrane);GO:0015031(protein transport);GO:0005768(endosome);GO:0070062(extracellular exosome);GO:0055037(recycling endosome);GO:0000149(SNARE binding);GO:0032456(endocytic recycling);GO:1990745(EARP complex);GO:1990745(EARP complex) | NA | NA | NA | VPS50, EARP/GARPII complex subunit [Source:HGNC Symbol;Acc:HGNC:25956] | 0.80 | 0.92 |
| ENSG00000004776 | HSPB6 | ENST00000004982 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0042803(protein homodimerization activity);GO:0005515(protein binding);GO:0005576(extracellular region);GO:0005212(structural constituent of eye lens);GO:0051082(unfolded protein binding);GO:0051087(chaperone binding);GO:0006937(regulation of muscle contraction);GO:0010667(negative regulation of cardiac muscle cell apoptotic process);GO:0045766(positive regulation of angiogenesis);GO:0061077(chaperone-mediated protein folding) | NA | NA | NA | heat shock protein family B (small) member 6 [Source:HGNC Symbol;Acc:HGNC:26511] | 0.08 | 0.35 |
| ENSG00000004777 | ARHGAP33 | ENST00000378944;ENST00000221905;ENST00000601474;ENST00000591438;ENST00000586918 | GO:0006810(transport);GO:0015031(protein transport);GO:0007165(signal transduction);GO:0043547(positive regulation of GTPase activity);GO:0005515(protein binding);GO:0005096(GTPase activator activity);GO:0035091(phosphatidylinositol binding);GO:0005829(cytosol);GO:0051056(regulation of small GTPase mediated signal transduction);GO:0019901(protein kinase binding);GO:0007264(small GTPase mediated signal transduction);GO:0043234(protein complex);GO:0005938(cell cortex);GO:0015629(actin cytoskeleton);GO:0005737(cytoplasm);GO:0005886(plasma membrane);GO:0005886(plasma membrane) | NA | NA | NA | Rho GTPase activating protein 33 [Source:HGNC Symbol;Acc:HGNC:23085] | 1.04 | 1.00 |
| ENSG00000004779 | NDUFAB1 | ENST00000007516;ENST00000484769;ENST00000570319;ENST00000567761 | GO:0005654(nucleoplasm);GO:0055114(oxidation-reduction process);GO:0005739(mitochondrion);GO:0006629(lipid metabolic process);GO:0006631(fatty acid metabolic process);GO:0006633(fatty acid biosynthetic process);GO:0005759(mitochondrial matrix);GO:0070469(respiratory chain);GO:0009249(protein lipoylation);GO:0005509(calcium ion binding);GO:0031966(mitochondrial membrane);GO:0005515(protein binding);GO:0032981(mitochondrial respiratory chain complex I assembly);GO:0005829(cytosol);GO:0034641(cellular nitrogen compound metabolic process);GO:0008137(NADH dehydrogenase (ubiquinone) activity);GO:0006120(mitochondrial electron transport, NADH to ubiquinone);GO:0005747(mitochondrial respiratory chain complex I);GO:0005743(mitochondrial inner membrane);GO:0000035(acyl binding);GO:0000036(ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process);GO:0005504(fatty acid binding);GO:0031177(phosphopantetheine binding);GO:0006635(fatty acid beta-oxidation);GO:0009245(lipid A biosynthetic process);GO:0009245(lipid A biosynthetic process) | 00190(Oxidative phosphorylation);04723(Retrograde endocannabinoid signaling);05010(Alzheimer's disease);05012(Parkinson's disease);05016(Huntington's disease);04932(Non-alcoholic fatty liver disease (NAFLD));04932(Non-alcoholic fatty liver disease (NAFLD)) | K03955 | NA | NADH:ubiquinone oxidoreductase subunit AB1 [Source:HGNC Symbol;Acc:HGNC:7694] | 11.424051114375 | 9.47 |
| ENSG00000004799 | PDK4 | ENST00000005178 | GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0016740(transferase activity);GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0006468(protein phosphorylation);GO:0005739(mitochondrion);GO:0005759(mitochondrial matrix);GO:0005975(carbohydrate metabolic process);GO:0006885(regulation of pH);GO:0008286(insulin receptor signaling pathway);GO:0010510(regulation of acetyl-CoA biosynthetic process from pyruvate);GO:0010565(regulation of cellular ketone metabolic process);GO:0010906(regulation of glucose metabolic process);GO:0042304(regulation of fatty acid biosynthetic process);GO:0042593(glucose homeostasis);GO:0042594(response to starvation);GO:0045124(regulation of bone resorption);GO:0046320(regulation of fatty acid oxidation);GO:0005743(mitochondrial inner membrane);GO:0009267(cellular response to starvation);GO:0006006(glucose metabolic process);GO:0004672(protein kinase activity);GO:0071398(cellular response to fatty acid);GO:0072593(reactive oxygen species metabolic process);GO:2000811(negative regulation of anoikis);GO:0004740(pyruvate dehydrogenase (acetyl-transferring) kinase activity) | NA | NA | NA | pyruvate dehydrogenase kinase 4 [Source:HGNC Symbol;Acc:HGNC:8812] | 2.20 | 2.50 |
| ENSG00000004838 | ZMYND10 | ENST00000231749;ENST00000475688 | GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0005815(microtubule organizing center);GO:0036159(inner dynein arm assembly);GO:0044458(motile cilium assembly);GO:0034451(centriolar satellite);GO:0036158(outer dynein arm assembly);GO:0036158(outer dynein arm assembly) | NA | NA | NA | zinc finger MYND-type containing 10 [Source:HGNC Symbol;Acc:HGNC:19412] | 0.04 | 0.04 |
| ENSG00000004846 | ABCB5 | ENST00000404938 | GO:0006810(transport);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0055085(transmembrane transport);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0005886(plasma membrane);GO:0030154(cell differentiation);GO:0042626(ATPase activity, coupled to transmembrane movement of substances);GO:0016887(ATPase activity);GO:0005887(integral component of plasma membrane);GO:0015562(efflux transmembrane transporter activity);GO:0006855(drug transmembrane transport);GO:0042391(regulation of membrane potential);GO:0008559(xenobiotic-transporting ATPase activity);GO:0042908(xenobiotic transport);GO:0048058(compound eye corneal lens development) | 02010(ABC transporters) | K05660 | NA | ATP binding cassette subfamily B member 5 [Source:HGNC Symbol;Acc:HGNC:46] | 0 | 0.00 |
| ENSG00000004864 | SLC25A13 | ENST00000265631;ENST00000416240;ENST00000490072;ENST00000492869 | GO:0006810(transport);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0055085(transmembrane transport);GO:0046872(metal ion binding);GO:0005739(mitochondrion);GO:0005743(mitochondrial inner membrane);GO:0005509(calcium ion binding);GO:0089711(L-glutamate transmembrane transport);GO:0005887(integral component of plasma membrane);GO:0006754(ATP biosynthetic process);GO:0005313(L-glutamate transmembrane transporter activity);GO:0006094(gluconeogenesis);GO:0045333(cellular respiration);GO:0005215(transporter activity);GO:0051592(response to calcium ion);GO:0015813(L-glutamate transport);GO:0015172(acidic amino acid transmembrane transporter activity);GO:0015183(L-aspartate transmembrane transporter activity);GO:0015810(aspartate transport);GO:0043490(malate-aspartate shuttle);GO:0089712(L-aspartate transmembrane transport);GO:0089712(L-aspartate transmembrane transport) | NA | NA | NA | solute carrier family 25 member 13 [Source:HGNC Symbol;Acc:HGNC:10983] | 11.3813599245283 | 6.79 |
| ENSG00000004866 | ST7 | ENST00000265437;ENST00000434836;ENST00000393443;ENST00000543837 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0016021(integral component of membrane) | NA | NA | NA | suppression of tumorigenicity 7 [Source:HGNC Symbol;Acc:HGNC:11351] | 1.81 | 2.28 |
| ENSG00000004897 | CDC27 | ENST00000066544 | GO:0005634(nucleus);GO:0005654(nucleoplasm);GO:0016567(protein ubiquitination);GO:0005515(protein binding);GO:0005737(cytoplasm);GO:0005829(cytosol);GO:0031145(anaphase-promoting complex-dependent catabolic process);GO:0043161(proteasome-mediated ubiquitin-dependent protein catabolic process);GO:0051437(positive regulation of ubiquitin-protein ligase activity involved in regulation of mitotic cell cycle transition);GO:0051436(negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle);GO:0008283(cell proliferation);GO:0042787(protein ubiquitination involved in ubiquitin-dependent protein catabolic process);GO:0051439(regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle);GO:0070979(protein K11-linked ubiquitination);GO:0005680(anaphase-promoting complex);GO:0019903(protein phosphatase binding);GO:0005813(centrosome);GO:0005819(spindle);GO:0005876(spindle microtubule);GO:0007091(metaphase/anaphase transition of mitotic cell cycle);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0051301(cell division) | 04120(Ubiquitin mediated proteolysis);04110(Cell cycle);04114(Oocyte meiosis);04914(Progesterone-mediated oocyte maturation);05166(HTLV-I infection) | K03350 | NA | cell division cycle 27 [Source:HGNC Symbol;Acc:HGNC:1728] | 4.95 | 5.27 |
| ENSG00000004948 | CALCR | ENST00000359558 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0004930(G-protein coupled receptor activity);GO:0007186(G-protein coupled receptor signaling pathway);GO:0005886(plasma membrane);GO:0004871(signal transducer activity);GO:0007165(signal transduction);GO:0004888(transmembrane signaling receptor activity);GO:0007166(cell surface receptor signaling pathway);GO:0004948(calcitonin receptor activity);GO:0015031(protein transport);GO:0005887(integral component of plasma membrane);GO:0005515(protein binding);GO:0004872(receptor activity);GO:0010628(positive regulation of gene expression);GO:0008565(protein transporter activity);GO:0007189(adenylate cyclase-activating G-protein coupled receptor signaling pathway);GO:0097643(amylin receptor activity);GO:0030816(positive regulation of cAMP metabolic process);GO:0030819(positive regulation of cAMP biosynthetic process);GO:0031623(receptor internalization);GO:0072659(protein localization to plasma membrane);GO:0097647(amylin receptor signaling pathway);GO:1903440(amylin receptor complex);GO:0010739(positive regulation of protein kinase A signaling);GO:0010942(positive regulation of cell death);GO:0033138(positive regulation of peptidyl-serine phosphorylation);GO:0051897(positive regulation of protein kinase B signaling);GO:0070374(positive regulation of ERK1 and ERK2 cascade);GO:1905665(positive regulation of calcium ion import across plasma membrane);GO:0045762(positive regulation of adenylate cyclase activity);GO:0051384(response to glucocorticoid);GO:0032841(calcitonin binding);GO:0007204(positive regulation of cytosolic calcium ion concentration);GO:0030279(negative regulation of ossification);GO:0030316(osteoclast differentiation);GO:0043488(regulation of mRNA stability);GO:0001669(acrosomal vesicle);GO:0005929(cilium) | 04080(Neuroactive ligand-receptor interaction);04380(Osteoclast differentiation) | K04576 | NA | calcitonin receptor [Source:HGNC Symbol;Acc:HGNC:1440] | 0 | 0.02 |
| ENSG00000004961 | HCCS | ENST00000321143 | GO:0016020(membrane);GO:0046872(metal ion binding);GO:0005739(mitochondrion);GO:0005743(mitochondrial inner membrane);GO:0016829(lyase activity);GO:0004408(holocytochrome-c synthase activity);GO:0009887(animal organ morphogenesis);GO:0055114(oxidation-reduction process);GO:0018063(cytochrome c-heme linkage) | 00860(Porphyrin and chlorophyll metabolism) | K01764 | EC:4.4.1.17 | holocytochrome c synthase [Source:HGNC Symbol;Acc:HGNC:4837] | 10.5759736111349 | 11.5431174154176 |
| ENSG00000004975 | DVL2 | ENST00000005340;ENST00000576840;ENST00000571745;ENST00000574143;ENST00000572285 | GO:0051091(positive regulation of sequence-specific DNA binding transcription factor activity);GO:0005737(cytoplasm);GO:0001934(positive regulation of protein phosphorylation);GO:0003151(outflow tract morphogenesis);GO:0043507(positive regulation of JUN kinase activity);GO:0005634(nucleus);GO:0016020(membrane);GO:0005654(nucleoplasm);GO:0035556(intracellular signal transduction);GO:0005829(cytosol);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0016235(aggresome);GO:0016604(nuclear body);GO:0007275(multicellular organism development);GO:0016055(Wnt signaling pathway);GO:0031410(cytoplasmic vesicle);GO:0042802(identical protein binding);GO:0006366(transcription from RNA polymerase II promoter);GO:0061024(membrane organization);GO:0060071(Wnt signaling pathway, planar cell polarity pathway);GO:0090090(negative regulation of canonical Wnt signaling pathway);GO:0045893(positive regulation of transcription, DNA-templated);GO:0060070(canonical Wnt signaling pathway);GO:0005109(frizzled binding);GO:0090179(planar cell polarity pathway involved in neural tube closure);GO:1904886(beta-catenin destruction complex disassembly);GO:0019901(protein kinase binding);GO:0019904(protein domain specific binding);GO:0030674(protein binding, bridging);GO:0043621(protein self-association);GO:0048365(Rac GTPase binding);GO:0001843(neural tube closure);GO:0003007(heart morphogenesis);GO:0007379(segment specification);GO:0007507(heart development);GO:0022007(convergent extension involved in neural plate elongation);GO:0034613(cellular protein localization);GO:0035282(segmentation);GO:0035329(hippo signaling);GO:0035567(non-canonical Wnt signaling pathway);GO:0043547(positive regulation of GTPase activity);GO:0044340(canonical Wnt signaling pathway involved in regulation of cell proliferation);GO:0051259(protein oligomerization);GO:0060029(convergent extension involved in organogenesis);GO:0061098(positive regulation of protein tyrosine kinase activity);GO:0090103(cochlea morphogenesis);GO:0090263(positive regulation of canonical Wnt signaling pathway);GO:0016328(lateral plasma membrane);GO:0030136(clathrin-coated vesicle);GO:0045177(apical part of cell);GO:0045334(clathrin-coated endocytic vesicle);GO:0005622(intracellular);GO:0005622(intracellular) | 04310(Wnt signaling pathway);04330(Notch signaling pathway);04390(Hippo signaling pathway);04150(mTOR signaling pathway);04550(Signaling pathways regulating pluripotency of stem cells);04916(Melanogenesis);05200(Pathways in cancer);05225(Hepatocellular carcinoma);05226(Gastric cancer);05217(Basal cell carcinoma);05224(Breast cancer);05166(HTLV-I infection);05165(Human papillomavirus infection);05165(Human papillomavirus infection) | K02353 | NA | dishevelled segment polarity protein 2 [Source:HGNC Symbol;Acc:HGNC:3086] | 6.85 | 6.68 |
| ENSG00000005001 | PRSS22 | ENST00000161006;ENST00000576381;ENST00000572061;ENST00000570950 | GO:0016787(hydrolase activity);GO:0008233(peptidase activity);GO:0006508(proteolysis);GO:0005576(extracellular region);GO:0004252(serine-type endopeptidase activity);GO:0008236(serine-type peptidase activity);GO:0046658(anchored component of plasma membrane);GO:0019897(extrinsic component of plasma membrane);GO:0019897(extrinsic component of plasma membrane) | NA | NA | NA | protease, serine 22 [Source:HGNC Symbol;Acc:HGNC:14368] | 0.07 | 0.01 |
| ENSG00000005007 | UPF1 | ENST00000262803;ENST00000601981;ENST00000600012;ENST00000598209;ENST00000600310;ENST00000598471;ENST00000596842 | GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0008270(zinc ion binding);GO:0046872(metal ion binding);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0005654(nucleoplasm);GO:0016787(hydrolase activity);GO:0003677(DNA binding);GO:0004386(helicase activity);GO:0000184(nuclear-transcribed mRNA catabolic process, nonsense-mediated decay);GO:0005515(protein binding);GO:0000932(P-body);GO:0000956(nuclear-transcribed mRNA catabolic process);GO:0005829(cytosol);GO:0071222(cellular response to lipopolysaccharide);GO:0061158(3'-UTR-mediated mRNA destabilization);GO:0000784(nuclear chromosome, telomeric region);GO:0071347(cellular response to interleukin-1);GO:0061014(positive regulation of mRNA catabolic process);GO:0035145(exon-exon junction complex);GO:0004004(ATP-dependent RNA helicase activity);GO:0071044(histone mRNA catabolic process);GO:0042162(telomeric DNA binding);GO:0032204(regulation of telomere maintenance);GO:0044770(cell cycle phase transition);GO:0044530(supraspliceosomal complex);GO:0006260(DNA replication);GO:0000294(nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay);GO:0006406(mRNA export from nucleus);GO:0006449(regulation of translational termination);GO:0000785(chromatin);GO:0003682(chromatin binding);GO:0006281(DNA repair);GO:0032201(telomere maintenance via semi-conservative replication);GO:0009048(dosage compensation by inactivation of X chromosome);GO:0009048(dosage compensation by inactivation of X chromosome) | 03013(RNA transport);03015(mRNA surveillance pathway);03015(mRNA surveillance pathway) | K14326 | EC:3.6.4.- | UPF1, RNA helicase and ATPase [Source:HGNC Symbol;Acc:HGNC:9962] | 13.3365730255814 | 12.5983437909302 |
| ENSG00000005020 | SKAP2 | ENST00000345317;ENST00000468712;ENST00000432747;ENST00000481204;ENST00000487720 | GO:0005737(cytoplasm);GO:0009967(positive regulation of signal transduction);GO:0005515(protein binding);GO:0005829(cytosol);GO:0005886(plasma membrane);GO:0042113(B cell activation);GO:0007165(signal transduction);GO:0006461(protein complex assembly);GO:0005070(SH3/SH2 adaptor activity);GO:0002757(immune response-activating signal transduction);GO:0008285(negative regulation of cell proliferation);GO:0008285(negative regulation of cell proliferation) | NA | NA | NA | src kinase associated phosphoprotein 2 [Source:HGNC Symbol;Acc:HGNC:15687] | 3.55 | 3.52 |
| ENSG00000005022 | SLC25A5 | ENST00000317881;ENST00000475354;ENST00000460013 | GO:0003723(RNA binding);GO:0006810(transport);GO:0005634(nucleus);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005215(transporter activity);GO:0055085(transmembrane transport);GO:0005739(mitochondrion);GO:0005743(mitochondrial inner membrane);GO:0005515(protein binding);GO:0007059(chromosome segregation);GO:0070062(extracellular exosome);GO:0016032(viral process);GO:0005887(integral component of plasma membrane);GO:0031012(extracellular matrix);GO:0008284(positive regulation of cell proliferation);GO:1901029(negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway);GO:0042645(mitochondrial nucleoid);GO:0050796(regulation of insulin secretion);GO:0031625(ubiquitin protein ligase binding);GO:0015866(ADP transport);GO:0071817(MMXD complex);GO:0005471(ATP:ADP antiporter activity);GO:0015207(adenine transmembrane transporter activity);GO:0015853(adenine transport);GO:0015867(ATP transport);GO:1990830(cellular response to leukemia inhibitory factor);GO:0043209(myelin sheath);GO:0045121(membrane raft);GO:0045121(membrane raft) | 04020(Calcium signaling pathway);04022(cGMP - PKG signaling pathway);04217(Necroptosis);04218(Cellular senescence);05012(Parkinson's disease);05016(Huntington's disease);05166(HTLV-I infection);05166(HTLV-I infection) | K05863 | NA | solute carrier family 25 member 5 [Source:HGNC Symbol;Acc:HGNC:10991] | 90.068431388 | 91.6869279393334 |
| ENSG00000005059 | MCUB | ENST00000394650;ENST00000452915 | GO:0006810(transport);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005743(mitochondrial inner membrane);GO:0005739(mitochondrion);GO:0006811(ion transport);GO:0031305(integral component of mitochondrial inner membrane);GO:0006816(calcium ion transport);GO:0065009(regulation of molecular function);GO:0031224(intrinsic component of membrane);GO:0006851(mitochondrial calcium ion transmembrane transport);GO:0051560(mitochondrial calcium ion homeostasis);GO:0019855(calcium channel inhibitor activity);GO:0034704(calcium channel complex);GO:1990246(uniplex complex);GO:0036444(mitochondrial calcium uptake);GO:0036444(mitochondrial calcium uptake) | NA | NA | NA | mitochondrial calcium uniporter dominant negative beta subunit [Source:HGNC Symbol;Acc:HGNC:26076] | 1.63 | 2.60 |
| ENSG00000005075 | POLR2J | ENST00000292614;ENST00000393794 | GO:0005634(nucleus);GO:0046983(protein dimerization activity);GO:0003677(DNA binding);GO:0006351(transcription, DNA-templated);GO:0005515(protein binding);GO:0003899(DNA-directed 5'-3' RNA polymerase activity);GO:0005654(nucleoplasm);GO:0006366(transcription from RNA polymerase II promoter);GO:0006368(transcription elongation from RNA polymerase II promoter);GO:0006370(7-methylguanosine mRNA capping);GO:0050434(positive regulation of viral transcription);GO:0006367(transcription initiation from RNA polymerase II promoter);GO:0042795(snRNA transcription from RNA polymerase II promoter);GO:0000398(mRNA splicing, via spliceosome);GO:0060964(regulation of gene silencing by miRNA);GO:0006283(transcription-coupled nucleotide-excision repair);GO:0008543(fibroblast growth factor receptor signaling pathway);GO:0016070(RNA metabolic process);GO:0035019(somatic stem cell population maintenance);GO:0005665(DNA-directed RNA polymerase II, core complex);GO:0001055(RNA polymerase II activity);GO:0030275(LRR domain binding);GO:0030275(LRR domain binding) | 00230(Purine metabolism);00240(Pyrimidine metabolism);03020(RNA polymerase);05016(Huntington's disease);05169(Epstein-Barr virus infection);05169(Epstein-Barr virus infection) | K03008 | NA | RNA polymerase II subunit J [Source:HGNC Symbol;Acc:HGNC:9197] | 18.7005748103321 | 14.9102474273063 |
| ENSG00000005100 | DHX33 | ENST00000225296;ENST00000575153 | GO:0003676(nucleic acid binding);GO:0003723(RNA binding);GO:0005634(nucleus);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0005730(nucleolus);GO:0016787(hydrolase activity);GO:0004386(helicase activity);GO:0005654(nucleoplasm);GO:0005737(cytoplasm);GO:0004004(ATP-dependent RNA helicase activity);GO:0000182(rDNA binding);GO:0045943(positive regulation of transcription from RNA polymerase I promoter);GO:0006396(RNA processing);GO:0033613(activating transcription factor binding);GO:0033613(activating transcription factor binding) | 04621(NOD-like receptor signaling pathway);04621(NOD-like receptor signaling pathway) | K17820 | EC:3.6.4.13 | DEAH-box helicase 33 [Source:HGNC Symbol;Acc:HGNC:16718] | 9.33 | 9.26 |
| ENSG00000005108 | THSD7A | ENST00000423059 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005886(plasma membrane);GO:0005576(extracellular region);GO:0030154(cell differentiation);GO:0070062(extracellular exosome);GO:0001525(angiogenesis) | NA | NA | NA | thrombospondin type 1 domain containing 7A [Source:HGNC Symbol;Acc:HGNC:22207] | 0.06 | 0.10 |
| ENSG00000005156 | LIG3 | ENST00000378526;ENST00000262327;ENST00000590181;ENST00000586407;ENST00000586058;ENST00000593099 | GO:0005634(nucleus);GO:0008270(zinc ion binding);GO:0046872(metal ion binding);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0005654(nucleoplasm);GO:0003677(DNA binding);GO:0051301(cell division);GO:0005739(mitochondrion);GO:0006281(DNA repair);GO:0016874(ligase activity);GO:0005515(protein binding);GO:0006310(DNA recombination);GO:0006974(cellular response to DNA damage stimulus);GO:0007049(cell cycle);GO:0006260(DNA replication);GO:0003909(DNA ligase activity);GO:0003910(DNA ligase (ATP) activity);GO:0051103(DNA ligation involved in DNA repair);GO:0071897(DNA biosynthetic process);GO:0006283(transcription-coupled nucleotide-excision repair);GO:0000724(double-strand break repair via homologous recombination);GO:0006288(base-excision repair, DNA ligation);GO:0006297(nucleotide-excision repair, DNA gap filling);GO:0006302(double-strand break repair);GO:0090298(negative regulation of mitochondrial DNA replication);GO:0006266(DNA ligation);GO:0006266(DNA ligation) | 03410(Base excision repair);03410(Base excision repair) | K10776 | EC:6.5.1.1 | DNA ligase 3 [Source:HGNC Symbol;Acc:HGNC:6600] | 2.59 | 2.60 |
| ENSG00000005175 | RPAP3 | ENST00000005386;ENST00000548211;ENST00000551293 | GO:0005515(protein binding);GO:0097255(R2TP complex);GO:0097255(R2TP complex) | NA | NA | NA | RNA polymerase II associated protein 3 [Source:HGNC Symbol;Acc:HGNC:26151] | 2.26 | 2.54 |
| ENSG00000005187 | ACSM3 | ENST00000501740;ENST00000614721;ENST00000289416;ENST00000562251;ENST00000567711 | GO:0042632(cholesterol homeostasis);GO:0046872(metal ion binding);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0003824(catalytic activity);GO:0006629(lipid metabolic process);GO:0008152(metabolic process);GO:0006631(fatty acid metabolic process);GO:0005739(mitochondrion);GO:0016874(ligase activity);GO:0005759(mitochondrial matrix);GO:0003674(molecular_function);GO:0005575(cellular_component);GO:0008217(regulation of blood pressure);GO:0003996(acyl-CoA ligase activity);GO:0004321(fatty-acyl-CoA synthase activity);GO:0015645(fatty acid ligase activity);GO:0047760(butyrate-CoA ligase activity);GO:0006633(fatty acid biosynthetic process);GO:0006637(acyl-CoA metabolic process);GO:0006637(acyl-CoA metabolic process) | 00650(Butanoate metabolism);00650(Butanoate metabolism) | K01896 | EC:6.2.1.2 | acyl-CoA synthetase medium chain family member 3 [Source:HGNC Symbol;Acc:HGNC:10522] | 0.70 | 0.82 |
| ENSG00000005189 | REXO5 | ENST00000566993;ENST00000261377;ENST00000563068;ENST00000563617;ENST00000563045 | GO:0003676(nucleic acid binding);GO:0003723(RNA binding);GO:0016787(hydrolase activity);GO:0090305(nucleic acid phosphodiester bond hydrolysis);GO:0004527(exonuclease activity);GO:0004518(nuclease activity);GO:0070062(extracellular exosome);GO:0005730(nucleolus);GO:0005730(nucleolus) | 03008(Ribosome biogenesis in eukaryotes);03008(Ribosome biogenesis in eukaryotes) | K14570 | EC:3.1.-.- | RNA exonuclease 5 [Source:HGNC Symbol;Acc:HGNC:24661] | 1.99 | 2.09 |
| ENSG00000005194 | CIAPIN1 | ENST00000394391;ENST00000563561;ENST00000566284 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0005654(nucleoplasm);GO:0008168(methyltransferase activity);GO:0005739(mitochondrion);GO:0008152(metabolic process);GO:0005515(protein binding);GO:0006915(apoptotic process);GO:0032259(methylation);GO:0051536(iron-sulfur cluster binding);GO:0005730(nucleolus);GO:0016226(iron-sulfur cluster assembly);GO:0051537(2 iron, 2 sulfur cluster binding);GO:0005758(mitochondrial intermembrane space);GO:0043066(negative regulation of apoptotic process);GO:0030097(hemopoiesis);GO:0030097(hemopoiesis) | NA | NA | NA | cytokine induced apoptosis inhibitor 1 [Source:HGNC Symbol;Acc:HGNC:28050] | 6.63 | 5.13 |
| ENSG00000005206 | SPPL2B | ENST00000613503;ENST00000586332;ENST00000614794;ENST00000586111;ENST00000593243 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005654(nucleoplasm);GO:0016787(hydrolase activity);GO:0005794(Golgi apparatus);GO:0005765(lysosomal membrane);GO:0005886(plasma membrane);GO:0005813(centrosome);GO:0042803(protein homodimerization activity);GO:0015629(actin cytoskeleton);GO:0008233(peptidase activity);GO:0006508(proteolysis);GO:0000139(Golgi membrane);GO:0004190(aspartic-type endopeptidase activity);GO:0005764(lysosome);GO:0031293(membrane protein intracellular domain proteolysis);GO:0005515(protein binding);GO:0005768(endosome);GO:0010008(endosome membrane);GO:0030660(Golgi-associated vesicle membrane);GO:0006509(membrane protein ectodomain proteolysis);GO:0010803(regulation of tumor necrosis factor-mediated signaling pathway);GO:0033619(membrane protein proteolysis);GO:0050776(regulation of immune response);GO:0042500(aspartic endopeptidase activity, intramembrane cleaving);GO:0071458(integral component of cytoplasmic side of endoplasmic reticulum membrane);GO:0071556(integral component of lumenal side of endoplasmic reticulum membrane);GO:0071556(integral component of lumenal side of endoplasmic reticulum membrane) | NA | NA | NA | signal peptide peptidase like 2B [Source:HGNC Symbol;Acc:HGNC:30627] | 2.55 | 2.23 |
| ENSG00000005238 | FAM214B | ENST00000378566;ENST00000603301;ENST00000378561;ENST00000605392 | GO:0005515(protein binding);GO:0005634(nucleus);GO:0005634(nucleus) | NA | NA | NA | family with sequence similarity 214 member B [Source:HGNC Symbol;Acc:HGNC:25666] | 1.66 | 2.09 |
| ENSG00000005243 | COPZ2 | ENST00000580409;ENST00000621465;ENST00000579263;ENST00000580174 | GO:0006810(transport);GO:0015031(protein transport);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0005794(Golgi apparatus);GO:0000139(Golgi membrane);GO:0031410(cytoplasmic vesicle);GO:0006886(intracellular protein transport);GO:0016192(vesicle-mediated transport);GO:0030663(COPI-coated vesicle membrane);GO:0005789(endoplasmic reticulum membrane);GO:0005829(cytosol);GO:0006890(retrograde vesicle-mediated transport, Golgi to ER);GO:0006888(ER to Golgi vesicle-mediated transport);GO:0030133(transport vesicle);GO:0030126(COPI vesicle coat);GO:0033116(endoplasmic reticulum-Golgi intermediate compartment membrane);GO:0005801(cis-Golgi network);GO:0005801(cis-Golgi network) | NA | NA | NA | coatomer protein complex subunit zeta 2 [Source:HGNC Symbol;Acc:HGNC:19356] | 2.54 | 3.04 |
| ENSG00000005249 | PRKAR2B | ENST00000265717 | GO:2000480(negative regulation of cAMP-dependent protein kinase activity);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0000166(nucleotide binding);GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0008603(cAMP-dependent protein kinase regulator activity);GO:0030552(cAMP binding);GO:0001932(regulation of protein phosphorylation);GO:0005952(cAMP-dependent protein kinase complex);GO:0019901(protein kinase binding);GO:0019904(protein domain specific binding);GO:0006631(fatty acid metabolic process);GO:0007612(learning);GO:0045859(regulation of protein kinase activity);GO:0097332(response to antipsychotic drug);GO:0097338(response to clozapine);GO:0005743(mitochondrial inner membrane);GO:0030425(dendrite);GO:0043025(neuronal cell body);GO:0043197(dendritic spine);GO:0043198(dendritic shaft);GO:0045121(membrane raft);GO:0048471(perinuclear region of cytoplasm);GO:0097546(ciliary base);GO:0098794(postsynapse);GO:0005813(centrosome);GO:0005829(cytosol);GO:0070062(extracellular exosome);GO:0031625(ubiquitin protein ligase binding);GO:0000086(G2/M transition of mitotic cell cycle);GO:0010389(regulation of G2/M transition of mitotic cell cycle);GO:0097711(ciliary basal body docking);GO:0035556(intracellular signal transduction);GO:0007596(blood coagulation);GO:0003091(renal water homeostasis);GO:0034199(activation of protein kinase A activity);GO:0071377(cellular response to glucagon stimulus);GO:0004862(cAMP-dependent protein kinase inhibitor activity);GO:0034236(protein kinase A catalytic subunit binding) | 04910(Insulin signaling pathway) | K04739 | NA | protein kinase cAMP-dependent type II regulatory subunit beta [Source:HGNC Symbol;Acc:HGNC:9392] | 2.48 | 2.75 |
| ENSG00000005302 | MSL3 | ENST00000361672;ENST00000312196;ENST00000337339;ENST00000398527 | GO:0005634(nucleus);GO:0006355(regulation of transcription, DNA-templated);GO:0006351(transcription, DNA-templated);GO:0016569(covalent chromatin modification);GO:0003677(DNA binding);GO:0006325(chromatin organization);GO:0005654(nucleoplasm);GO:0006338(chromatin remodeling);GO:0035064(methylated histone binding);GO:0043984(histone H4-K16 acetylation);GO:0006342(chromatin silencing);GO:0016575(histone deacetylation);GO:0043968(histone H2A acetylation);GO:0035267(NuA4 histone acetyltransferase complex);GO:0072487(MSL complex);GO:0072487(MSL complex) | NA | NA | NA | MSL complex subunit 3 [Source:HGNC Symbol;Acc:HGNC:7370] | 0.78 | 0.52 |
| ENSG00000005339 | CREBBP | ENST00000262367;ENST00000573517;ENST00000635899;ENST00000636895 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0008270(zinc ion binding);GO:0046872(metal ion binding);GO:0005654(nucleoplasm);GO:0016740(transferase activity);GO:0003713(transcription coactivator activity);GO:0006355(regulation of transcription, DNA-templated);GO:0006351(transcription, DNA-templated);GO:0016746(transferase activity, transferring acyl groups);GO:0005515(protein binding);GO:0000123(histone acetyltransferase complex);GO:0007165(signal transduction);GO:0016604(nuclear body);GO:0048511(rhythmic process);GO:0004402(histone acetyltransferase activity);GO:0045893(positive regulation of transcription, DNA-templated);GO:0003712(transcription cofactor activity);GO:0016573(histone acetylation);GO:0031648(protein destabilization);GO:0002223(stimulatory C-type lectin receptor signaling pathway);GO:0061418(regulation of transcription from RNA polymerase II promoter in response to hypoxia);GO:0003682(chromatin binding);GO:0001666(response to hypoxia);GO:0016032(viral process);GO:0004871(signal transducer activity);GO:0008134(transcription factor binding);GO:0042981(regulation of apoptotic process);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:1904837(beta-catenin-TCF complex assembly);GO:0000790(nuclear chromatin);GO:0006367(transcription initiation from RNA polymerase II promoter);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0001085(RNA polymerase II transcription factor binding);GO:0006461(protein complex assembly);GO:0042592(homeostatic process);GO:0007219(Notch signaling pathway);GO:0042733(embryonic digit morphogenesis);GO:0001078(transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding);GO:0001102(RNA polymerase II activating transcription factor binding);GO:0001191(transcriptional repressor activity, RNA polymerase II transcription factor binding);GO:0032481(positive regulation of type I interferon production);GO:0002039(p53 binding);GO:0019216(regulation of lipid metabolic process);GO:1900034(regulation of cellular response to heat);GO:0003684(damaged DNA binding);GO:0000987(core promoter proximal region sequence-specific DNA binding);GO:0034644(cellular response to UV);GO:0045637(regulation of myeloid cell differentiation);GO:0001105(RNA polymerase II transcription coactivator activity);GO:0016407(acetyltransferase activity);GO:0006473(protein acetylation);GO:0034212(peptide N-acetyltransferase activity);GO:0018076(N-terminal peptidyl-lysine acetylation);GO:0043426(MRF binding);GO:0008589(regulation of smoothened signaling pathway);GO:0008589(regulation of smoothened signaling pathway) | 04310(Wnt signaling pathway);04330(Notch signaling pathway);04350(TGF-beta signaling pathway);04630(Jak-STAT signaling pathway);04066(HIF-1 signaling pathway);04068(FoxO signaling pathway);04024(cAMP signaling pathway);04110(Cell cycle);04520(Adherens junction);04922(Glucagon signaling pathway);04919(Thyroid hormone signaling pathway);04916(Melanogenesis);04720(Long-term potentiation);05200(Pathways in cancer);05206(MicroRNAs in cancer);05203(Viral carcinogenesis);05211(Renal cell carcinoma);05215(Prostate cancer);05016(Huntington's disease);05152(Tuberculosis);05166(HTLV-I infection);05164(Influenza A);05161(Hepatitis B);05168(Herpes simplex infection);05167(Kaposi's sarcoma-associated herpesvirus infection);05169(Epstein-Barr virus infection);05165(Human papillomavirus infection);05165(Human papillomavirus infection) | K04498 | EC:2.3.1.48 | CREB binding protein [Source:HGNC Symbol;Acc:HGNC:2348] | 4.92 | 4.95 |
| ENSG00000005379 | TSPOAP1 | ENST00000581692 | GO:0030156(benzodiazepine receptor binding);GO:0005737(cytoplasm);GO:0005739(mitochondrion);GO:0005515(protein binding);GO:0098793(presynapse);GO:0008150(biological_process);GO:0005829(cytosol);GO:0014047(glutamate secretion);GO:0007269(neurotransmitter secretion);GO:0006700(C21-steroid hormone biosynthetic process) | NA | NA | NA | TSPO associated protein 1 [Source:HGNC Symbol;Acc:HGNC:16831] | 0 | 0.01 |
| ENSG00000005421 | PON1 | ENST00000222381 | GO:0005615(extracellular space);GO:0046872(metal ion binding);GO:0016787(hydrolase activity);GO:0005576(extracellular region);GO:0016311(dephosphorylation);GO:0005509(calcium ion binding);GO:0004064(arylesterase activity);GO:0070062(extracellular exosome);GO:0072562(blood microparticle);GO:0034364(high-density lipoprotein particle);GO:0042803(protein homodimerization activity);GO:0051099(positive regulation of binding);GO:0019372(lipoxygenase pathway);GO:0005543(phospholipid binding);GO:0034366(spherical high-density lipoprotein particle);GO:0010875(positive regulation of cholesterol efflux);GO:0046470(phosphatidylcholine metabolic process);GO:0009605(response to external stimulus);GO:0009636(response to toxic substance);GO:0004063(aryldialkylphosphatase activity);GO:0102007(acyl-L-homoserine-lactone lactonohydrolase activity);GO:0006629(lipid metabolic process);GO:0008203(cholesterol metabolic process);GO:0019439(aromatic compound catabolic process);GO:0031667(response to nutrient levels);GO:0032411(positive regulation of transporter activity);GO:0046395(carboxylic acid catabolic process);GO:0046434(organophosphate catabolic process);GO:0070542(response to fatty acid);GO:1902617(response to fluoride);GO:0043231(intracellular membrane-bounded organelle) | NA | NA | NA | paraoxonase 1 [Source:HGNC Symbol;Acc:HGNC:9204] | 0.05 | 0.06 |
| ENSG00000005436 | GCFC2 | ENST00000321027 | GO:0005634(nucleus);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0006397(mRNA processing);GO:0008380(RNA splicing);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0006351(transcription, DNA-templated);GO:0005515(protein binding);GO:0005730(nucleolus);GO:0000978(RNA polymerase II core promoter proximal region sequence-specific DNA binding);GO:0001078(transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0045892(negative regulation of transcription, DNA-templated);GO:0000398(mRNA splicing, via spliceosome);GO:0071008(U2-type post-mRNA release spliceosomal complex);GO:0000245(spliceosomal complex assembly) | NA | NA | NA | GC-rich sequence DNA-binding factor 2 [Source:HGNC Symbol;Acc:HGNC:1317] | 0.83 | 0.78 |
| ENSG00000005448 | WDR54 | ENST00000461531;ENST00000465134;ENST00000468778 | NA | NA | NA | NA | WD repeat domain 54 [Source:HGNC Symbol;Acc:HGNC:25770] | 3.38 | 2.12 |
| ENSG00000005469 | CROT | ENST00000419147;ENST00000412227;ENST00000442291 | GO:0006810(transport);GO:0043231(intracellular membrane-bounded organelle);GO:0016740(transferase activity);GO:0016746(transferase activity, transferring acyl groups);GO:0006629(lipid metabolic process);GO:0006631(fatty acid metabolic process);GO:0005777(peroxisome);GO:0005102(receptor binding);GO:0005782(peroxisomal matrix);GO:0033540(fatty acid beta-oxidation using acyl-CoA oxidase);GO:0006635(fatty acid beta-oxidation);GO:0006091(generation of precursor metabolites and energy);GO:0009437(carnitine metabolic process);GO:0008458(carnitine O-octanoyltransferase activity);GO:0015908(fatty acid transport);GO:0015936(coenzyme A metabolic process);GO:0051791(medium-chain fatty acid metabolic process);GO:0005739(mitochondrion);GO:0005739(mitochondrion) | 04146(Peroxisome);04146(Peroxisome) | K05940 | EC:2.3.1.137 | carnitine O-octanoyltransferase [Source:HGNC Symbol;Acc:HGNC:2366] | 1.10 | 1.15 |
| ENSG00000005471 | ABCB4 | ENST00000265723 | GO:0006810(transport);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0055085(transmembrane transport);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0016787(hydrolase activity);GO:0005886(plasma membrane);GO:0005925(focal adhesion);GO:0015629(actin cytoskeleton);GO:0006869(lipid transport);GO:0031410(cytoplasmic vesicle);GO:0042626(ATPase activity, coupled to transmembrane movement of substances);GO:0016887(ATPase activity);GO:0045121(membrane raft);GO:0005515(protein binding);GO:0070062(extracellular exosome);GO:0005887(integral component of plasma membrane);GO:0016324(apical plasma membrane);GO:0030136(clathrin-coated vesicle);GO:0006629(lipid metabolic process);GO:0032782(bile acid secretion);GO:0019216(regulation of lipid metabolic process);GO:0045332(phospholipid translocation);GO:2001140(positive regulation of phospholipid transport);GO:0042493(response to drug);GO:0042908(xenobiotic transport);GO:0090554(phosphatidylcholine-translocating ATPase activity);GO:0008559(xenobiotic-transporting ATPase activity);GO:0005548(phospholipid transporter activity);GO:0008525(phosphatidylcholine transporter activity);GO:0032376(positive regulation of cholesterol transport);GO:0055088(lipid homeostasis);GO:0061092(positive regulation of phospholipid translocation);GO:1901557(response to fenofibrate);GO:1903413(cellular response to bile acid);GO:0001666(response to hypoxia);GO:0001890(placenta development);GO:0006855(drug transmembrane transport);GO:0007420(brain development);GO:0007595(lactation);GO:0007623(circadian rhythm);GO:0009914(hormone transport);GO:0010033(response to organic substance);GO:0010046(response to mycotoxin);GO:0014070(response to organic cyclic compound);GO:0014072(response to isoquinoline alkaloid);GO:0031667(response to nutrient levels);GO:0032355(response to estradiol);GO:0033189(response to vitamin A);GO:0033231(carbohydrate export);GO:0033280(response to vitamin D);GO:0035633(maintenance of permeability of blood-brain barrier);GO:0036146(cellular response to mycotoxin);GO:0043215(daunorubicin transport);GO:0043278(response to morphine);GO:0046618(drug export);GO:0046686(response to cadmium ion);GO:0050892(intestinal absorption);GO:0060548(negative regulation of cell death);GO:0060856(establishment of blood-brain barrier);GO:0071217(cellular response to external biotic stimulus);GO:0071236(cellular response to antibiotic);GO:0071312(cellular response to alkaloid);GO:0071392(cellular response to estradiol stimulus);GO:0071407(cellular response to organic cyclic compound);GO:0071475(cellular hyperosmotic salinity response);GO:0071548(response to dexamethasone);GO:0071549(cellular response to dexamethasone stimulus);GO:0097068(response to thyroxine);GO:0097327(response to antineoplastic agent);GO:1902065(response to L-glutamate);GO:1903416(response to glycoside);GO:1904478(regulation of intestinal absorption);GO:1905231(cellular response to borneol);GO:1905232(cellular response to L-glutamate);GO:1905233(response to codeine);GO:1905235(response to quercetin);GO:1905237(response to cyclosporin A);GO:1990962(drug transport across blood-brain barrier);GO:1990963(establishment of blood-retinal barrier);GO:2001025(positive regulation of response to drug);GO:0031526(brush border membrane);GO:0045177(apical part of cell);GO:0046581(intercellular canaliculus) | 02010(ABC transporters);04976(Bile secretion) | K05659 | EC:3.6.3.44 | ATP binding cassette subfamily B member 4 [Source:HGNC Symbol;Acc:HGNC:45] | 0.00 | 0.02 |
| ENSG00000005483 | KMT2E | ENST00000311117;ENST00000476671;ENST00000482560;ENST00000478079 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016607(nuclear speck);GO:0016020(membrane);GO:0046872(metal ion binding);GO:0008168(methyltransferase activity);GO:0016740(transferase activity);GO:0032259(methylation);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0016569(covalent chromatin modification);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0007049(cell cycle);GO:0030218(erythrocyte differentiation);GO:0018024(histone-lysine N-methyltransferase activity);GO:0034968(histone lysine methylation);GO:0005654(nucleoplasm);GO:0007050(cell cycle arrest);GO:0045652(regulation of megakaryocyte differentiation);GO:0042119(neutrophil activation);GO:0006306(DNA methylation);GO:0002446(neutrophil mediated immunity);GO:0002446(neutrophil mediated immunity) | 00310(Lysine degradation);00310(Lysine degradation) | K09189 | EC:2.1.1.43 | lysine methyltransferase 2E [Source:HGNC Symbol;Acc:HGNC:18541] | 3.11 | 3.54 |
| ENSG00000005486 | RHBDD2 | ENST00000006777;ENST00000318622;ENST00000476218;ENST00000467406 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005794(Golgi apparatus);GO:0005634(nucleus);GO:0005654(nucleoplasm);GO:0004252(serine-type endopeptidase activity);GO:0006508(proteolysis);GO:0000139(Golgi membrane);GO:0048471(perinuclear region of cytoplasm);GO:0048471(perinuclear region of cytoplasm) | NA | NA | NA | rhomboid domain containing 2 [Source:HGNC Symbol;Acc:HGNC:23082] | 26.9508722798034 | 24.1745259303564 |
| ENSG00000005513 | SOX8 | ENST00000566034 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0006355(regulation of transcription, DNA-templated);GO:0006351(transcription, DNA-templated);GO:0003677(DNA binding);GO:0000981(RNA polymerase II transcription factor activity, sequence-specific DNA binding);GO:0007165(signal transduction);GO:0045893(positive regulation of transcription, DNA-templated);GO:0007283(spermatogenesis);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0045444(fat cell differentiation);GO:0060612(adipose tissue development);GO:0008584(male gonad development);GO:0043066(negative regulation of apoptotic process);GO:0045892(negative regulation of transcription, DNA-templated);GO:0045662(negative regulation of myoblast differentiation);GO:0060041(retina development in camera-type eye);GO:0001701(in utero embryonic development);GO:0045165(cell fate commitment);GO:0001755(neural crest cell migration);GO:0001649(osteoblast differentiation);GO:0000979(RNA polymerase II core promoter sequence-specific DNA binding);GO:0090190(positive regulation of branching involved in ureteric bud morphogenesis);GO:0033690(positive regulation of osteoblast proliferation);GO:0090184(positive regulation of kidney development);GO:0048709(oligodendrocyte differentiation);GO:0048484(enteric nervous system development);GO:0060221(retinal rod cell differentiation);GO:0061138(morphogenesis of a branching epithelium);GO:0072034(renal vesicle induction);GO:0072289(metanephric nephron tubule formation);GO:0007422(peripheral nervous system development);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0008134(transcription factor binding);GO:0043565(sequence-specific DNA binding);GO:0046982(protein heterodimerization activity);GO:0010628(positive regulation of gene expression);GO:0010817(regulation of hormone levels);GO:0014015(positive regulation of gliogenesis);GO:0035914(skeletal muscle cell differentiation);GO:0046533(negative regulation of photoreceptor cell differentiation);GO:0048469(cell maturation);GO:0060009(Sertoli cell development);GO:0060018(astrocyte fate commitment);GO:0072197(ureter morphogenesis);GO:0044798(nuclear transcription factor complex) | NA | NA | NA | SRY-box 8 [Source:HGNC Symbol;Acc:HGNC:11203] | 0 | 0.02 |
| ENSG00000005700 | IBTK | ENST00000306270;ENST00000505222;ENST00000508381;ENST00000471036 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0005654(nucleoplasm);GO:0019901(protein kinase binding);GO:0061099(negative regulation of protein tyrosine kinase activity);GO:0001933(negative regulation of protein phosphorylation);GO:0030292(protein tyrosine kinase inhibitor activity);GO:0051209(release of sequestered calcium ion into cytosol);GO:0051209(release of sequestered calcium ion into cytosol) | NA | NA | NA | inhibitor of Bruton tyrosine kinase [Source:HGNC Symbol;Acc:HGNC:17853] | 2.79 | 3.02 |
| ENSG00000005801 | ZNF195 | ENST00000528796;ENST00000354599;ENST00000620374;ENST00000429541;ENST00000529789;ENST00000529228;ENST00000526540;ENST00000526598;ENST00000528636;ENST00000330692;ENST00000526501;ENST00000527386 | GO:0003676(nucleic acid binding);GO:0006355(regulation of transcription, DNA-templated);GO:0005622(intracellular);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0006351(transcription, DNA-templated);GO:0003677(DNA binding);GO:0008270(zinc ion binding);GO:0008270(zinc ion binding) | NA | NA | NA | zinc finger protein 195 [Source:HGNC Symbol;Acc:HGNC:12986] | 2.36 | 2.41 |
| ENSG00000005810 | MYCBP2 | ENST00000544440;ENST00000482517;ENST00000462987 | GO:0005634(nucleus);GO:0016020(membrane);GO:0046872(metal ion binding);GO:0016740(transferase activity);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0016567(protein ubiquitination);GO:0005515(protein binding);GO:0004842(ubiquitin-protein transferase activity);GO:0004842(ubiquitin-protein transferase activity) | NA | NA | NA | MYC binding protein 2, E3 ubiquitin protein ligase [Source:HGNC Symbol;Acc:HGNC:23386] | 3.56 | 4.16 |
Definition of table:
| term | Description |
|---|---|
| gene_id | Defines the ID of gene |
| gene_name | Defines the name of gene or gene symbol |
| transcript_id | Defines the ID of transcript |
| GO | Gene ontology term and description http://www.geneontology.org/ |
| KEGG | KEGG pathway annotation and description http://www.kegg.jp/kegg/kegg3a.html |
| KO_Entry | KO functional ortholog, KO groups identified by K numbers and, in metabolic maps |
| EC | enzyme |
| Description | Description of gene |
| FPKM.A | Normalization of gene expression level by FPKM value |
document location:summary6_transcript_expression/4_genes_fpkm_expression.xlsx
| t_id | class_code | chr | strand | start | end | t_name | num_exons | length | gene_id | gene_name | GO | KEGG | KO_ENTRY | EC | Description | cov.M14_OE | FPKM.M14_OE | cov.M14_NC | FPKM.M14_NC |
| 1 | NA | chr1 | + | 11869 | 14409 | ENST00000456328 | 3 | 1657 | ENSG00000223972 | DDX11L1 | NA | NA | NA | NA | DEAD/H-box helicase 11 like 1 [Source:HGNC Symbol;Acc:HGNC:37102] | 0.10 | 0.01 | 0.32 | 0.04 |
| 2 | NA | chr1 | + | 12010 | 13670 | ENST00000450305 | 6 | 632 | ENSG00000223972 | DDX11L1 | NA | NA | NA | NA | DEAD/H-box helicase 11 like 1 [Source:HGNC Symbol;Acc:HGNC:37102] | 0.05 | 0.01 | 0 | 0 |
| 3 | NA | chr1 | - | 14404 | 29570 | ENST00000488147 | 11 | 1351 | ENSG00000227232 | WASH7P | NA | NA | NA | NA | WAS protein family homolog 7 pseudogene [Source:HGNC Symbol;Acc:HGNC:38034] | 31.995941 | 3.86 | 25.793186 | 3.32 |
| 5 | NA | chr1 | + | 29554 | 31097 | ENST00000473358 | 3 | 712 | ENSG00000243485 | MIR1302-2HG | NA | NA | NA | NA | MIR1302-2 host gene [Source:HGNC Symbol;Acc:HGNC:52482] | 0 | 0 | 0.20 | 0.03 |
| 11 | NA | chr1 | + | 57598 | 64116 | ENST00000642116 | 3 | 1414 | ENSG00000240361 | OR4G11P | NA | NA | NA | NA | olfactory receptor family 4 subfamily G member 11 pseudogene [Source:HGNC Symbol;Acc:HGNC:31276] | 0 | 0 | 0.16 | 0.02 |
| 14 | NA | chr1 | + | 69055 | 70108 | ENST00000335137 | 1 | 1054 | ENSG00000186092 | OR4F5 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0004930(G-protein coupled receptor activity);GO:0007186(G-protein coupled receptor signaling pathway);GO:0005886(plasma membrane);GO:0004871(signal transducer activity);GO:0007165(signal transduction);GO:0004984(olfactory receptor activity);GO:0007608(sensory perception of smell);GO:0050896(response to stimulus);GO:0050911(detection of chemical stimulus involved in sensory perception of smell);GO:0004888(transmembrane signaling receptor activity);GO:0050907(detection of chemical stimulus involved in sensory perception) | 04740(Olfactory transduction) | K04257 | NA | olfactory receptor family 4 subfamily F member 5 [Source:HGNC Symbol;Acc:HGNC:14825] | 0 | 0 | 0.28 | 0.04 |
| 15 | NA | chr1 | - | 89295 | 120932 | ENST00000466430 | 4 | 2748 | ENSG00000238009 | AL627309.1 | NA | NA | NA | NA | NA | 0.08 | 0.01 | 0.68 | 0.09 |
| 18 | NA | chr1 | - | 110953 | 129173 | ENST00000471248 | 3 | 629 | ENSG00000238009 | AL627309.1 | NA | NA | NA | NA | NA | 0.23 | 0.03 | 0 | 0 |
| 21 | NA | chr1 | + | 131025 | 134836 | ENST00000442987 | 1 | 3812 | ENSG00000233750 | CICP27 | NA | NA | NA | NA | capicua transcriptional repressor pseudogene 27 [Source:HGNC Symbol;Acc:HGNC:48835] | 0.50 | 0.06 | 0.55 | 0.07 |
| 24 | NA | chr1 | - | 139790 | 140339 | ENST00000493797 | 2 | 323 | ENSG00000239906 | AL627309.2 | NA | NA | NA | NA | NA | 0.18 | 0.02 | 0.12 | 0.02 |
| 25 | NA | chr1 | - | 141474 | 149707 | ENST00000484859 | 2 | 4860 | ENSG00000241860 | AL627309.5 | NA | NA | NA | NA | NA | 0.14 | 0.02 | 0.03 | 0.00 |
| 27 | NA | chr1 | - | 146386 | 173862 | ENST00000466557 | 8 | 1301 | ENSG00000241860 | AL627309.5 | NA | NA | NA | NA | NA | 1.37 | 0.17 | 1.08 | 0.14 |
| 29 | NA | chr1 | + | 160446 | 161525 | ENST00000496488 | 2 | 457 | ENSG00000241599 | AL627309.4 | NA | NA | NA | NA | NA | 0 | 0 | 0.29 | 0.04 |
| 30 | NA | chr1 | - | 165889 | 168767 | ENST00000491962 | 3 | 278 | ENSG00000241860 | AL627309.5 | NA | NA | NA | NA | NA | 0.10 | 0.01 | 0 | 0 |
| 31 | NA | chr1 | + | 182696 | 184174 | ENST00000624431 | 5 | 570 | ENSG00000279928 | FO538757.2 | NA | NA | NA | NA | NA | 1.21 | 0.15 | 3.00 | 0.39 |
| 32 | NA | chr1 | - | 185217 | 195411 | ENST00000623083 | 10 | 1397 | ENSG00000279457 | FO538757.1 | NA | NA | NA | NA | NA | 24.990286 | 3.01 | 30.232634 | 3.89 |
| 35 | NA | chr1 | - | 257913 | 268816 | ENST00000448958 | 4 | 2250 | ENSG00000228463 | AP006222.1 | NA | NA | NA | NA | NA | 0 | 0 | 0.12 | 0.02 |
| 36 | NA | chr1 | - | 258144 | 359681 | ENST00000441866 | 4 | 2256 | ENSG00000228463 | AP006222.1 | NA | NA | NA | NA | NA | 0.13 | 0.02 | 0.07 | 0.01 |
| 39 | NA | chr1 | - | 263015 | 297502 | ENST00000424587 | 4 | 5603 | ENSG00000228463 | AP006222.1 | NA | NA | NA | NA | NA | 0.23 | 0.03 | 0.02 | 0.00 |
| 51 | NA | chr1 | - | 484832 | 495476 | ENST00000601814 | 3 | 635 | ENSG00000237094 | AL732372.2 | NA | NA | NA | NA | NA | 0 | 0 | 0.05 | 0.01 |
| 54 | NA | chr1 | + | 487101 | 489906 | ENST00000432723 | 2 | 2477 | ENSG00000233653 | CICP7 | NA | NA | NA | NA | capicua transcriptional repressor pseudogene 7 [Source:HGNC Symbol;Acc:HGNC:37756] | 0.28 | 0.03 | 0.39 | 0.05 |
| 55 | NA | chr1 | - | 491225 | 493241 | ENST00000514436 | 2 | 1239 | ENSG00000250575 | AL732372.3 | NA | NA | NA | NA | NA | 1.60 | 0.19 | 2.08 | 0.27 |
| 56 | NA | chr1 | - | 494382 | 496605 | ENST00000419160 | 2 | 547 | ENSG00000237094 | AL732372.2 | NA | NA | NA | NA | NA | 0 | 0 | 0.68 | 0.09 |
| 57 | NA | chr1 | - | 494464 | 502508 | ENST00000440038 | 5 | 793 | ENSG00000237094 | AL732372.2 | NA | NA | NA | NA | NA | 4.00 | 0.48 | 3.18 | 0.41 |
| 62 | NA | chr1 | - | 497205 | 502598 | ENST00000641845 | 6 | 571 | ENSG00000237094 | AL732372.2 | NA | NA | NA | NA | NA | 0 | 0 | 0.03 | 0.00 |
| 64 | NA | chr1 | - | 497240 | 499002 | ENST00000601486 | 4 | 696 | ENSG00000237094 | AL732372.2 | NA | NA | NA | NA | NA | 4.50 | 0.54 | 2.67 | 0.34 |
| 70 | NA | chr1 | - | 501604 | 517225 | ENST00000641063 | 5 | 473 | ENSG00000237094 | AL732372.2 | NA | NA | NA | NA | NA | 0.10 | 0.01 | 0.15 | 0.02 |
| 71 | NA | chr1 | - | 504470 | 514413 | ENST00000641049 | 2 | 519 | ENSG00000237094 | AL732372.2 | NA | NA | NA | NA | NA | 1.44 | 0.17 | 1.13 | 0.15 |
| 72 | NA | chr1 | - | 504865 | 522928 | ENST00000642124 | 4 | 456 | ENSG00000237094 | AL732372.2 | NA | NA | NA | NA | NA | 0.15 | 0.02 | 0.03 | 0.00 |
| 80 | NA | chr1 | - | 594198 | 631204 | ENST00000641296 | 6 | 1127 | ENSG00000230021 | AL669831.3 | NA | NA | NA | NA | NA | 0.49 | 0.06 | 0.23 | 0.03 |
| 81 | NA | chr1 | - | 594308 | 598551 | ENST00000357876 | 2 | 1702 | ENSG00000230021 | AL669831.3 | NA | NA | NA | NA | NA | 0 | 0 | 0.17 | 0.02 |
| 87 | NA | chr1 | + | 629640 | 630683 | ENST00000457540 | 1 | 1044 | ENSG00000225630 | MTND2P28 | NA | NA | NA | NA | mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 2 pseudogene 28 [Source:HGNC Symbol;Acc:HGNC:42129] | 2019.034546 | 243.43898 | 1723.17749 | 221.695282 |
| 88 | NA | chr1 | + | 631074 | 632616 | ENST00000414273 | 1 | 1543 | ENSG00000237973 | MTCO1P12 | NA | NA | NA | NA | mitochondrially encoded cytochrome c oxidase I pseudogene 12 [Source:HGNC Symbol;Acc:HGNC:52014] | 811.739441 | 97.873032 | 831.252197 | 106.944695 |
| 92 | NA | chr1 | + | 633696 | 634376 | ENST00000514057 | 1 | 681 | ENSG00000248527 | MTATP6P1 | NA | NA | NA | NA | mitochondrially encoded ATP synthase 6 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:44575] | 0 | 0 | 3385.140381 | 435.515015 |
| 93 | NA | chr1 | + | 634376 | 634922 | ENST00000416718 | 1 | 547 | ENSG00000198744 | MTCO3P12 | NA | NA | NA | NA | mitochondrially encoded cytochrome c oxidase III pseudogene 12 [Source:HGNC Symbol;Acc:HGNC:52042] | 116.72374 | 14.073611 | 0 | 0 |
| 97 | NA | chr1 | - | 711867 | 732212 | ENST00000414688 | 3 | 421 | ENSG00000230021 | AL669831.3 | NA | NA | NA | NA | NA | 3.99 | 0.48 | 3.17 | 0.41 |
| 98 | NA | chr1 | - | 720024 | 720206 | ENST00000636676 | 1 | 183 | ENSG00000230021 | AL669831.3 | NA | NA | NA | NA | NA | 0.47 | 0.06 | 0 | 0 |
| 100 | NA | chr1 | + | 722092 | 724903 | ENST00000440782 | 2 | 2455 | ENSG00000229376 | CICP3 | NA | NA | NA | NA | capicua transcriptional repressor pseudogene 3 [Source:HGNC Symbol;Acc:HGNC:37742] | 0.75 | 0.09 | 0.43 | 0.06 |
| 101 | NA | chr1 | - | 725885 | 778626 | ENST00000506640 | 16 | 6432 | ENSG00000228327 | AL669831.1 | NA | NA | NA | NA | NA | 7.15 | 0.86 | 7.05 | 0.91 |
| 105 | NA | chr1 | + | 778770 | 784690 | ENST00000434264 | 3 | 844 | ENSG00000237491 | AL669831.5 | NA | NA | NA | NA | NA | 0.43 | 0.05 | 0.70 | 0.09 |
| 107 | NA | chr1 | + | 778930 | 784655 | ENST00000589899 | 4 | 809 | ENSG00000237491 | AL669831.5 | NA | NA | NA | NA | NA | 0.19 | 0.02 | 0.23 | 0.03 |
| 121 | NA | chr1 | + | 784396 | 807321 | ENST00000586288 | 7 | 1040 | ENSG00000237491 | AL669831.5 | NA | NA | NA | NA | NA | 0.08 | 0.01 | 0.43 | 0.06 |
| 122 | NA | chr1 | + | 785800 | 787672 | ENST00000591702 | 1 | 1873 | ENSG00000237491 | AL669831.5 | NA | NA | NA | NA | NA | 0.16 | 0.02 | 0.39 | 0.05 |
| 123 | NA | chr1 | - | 800879 | 817712 | ENST00000447500 | 5 | 872 | ENSG00000230092 | AL669831.4 | NA | NA | NA | NA | NA | 2.15 | 0.26 | 1.74 | 0.22 |
| 125 | NA | chr1 | + | 804932 | 810060 | ENST00000412115 | 3 | 581 | ENSG00000237491 | AL669831.5 | NA | NA | NA | NA | NA | 0.22 | 0.03 | 0.61 | 0.08 |
| 127 | NA | chr1 | + | 817371 | 819837 | ENST00000326734 | 2 | 1947 | ENSG00000177757 | FAM87B | NA | NA | NA | NA | family with sequence similarity 87 member B [Source:HGNC Symbol;Acc:HGNC:32236] | 0.13 | 0.02 | 0.15 | 0.02 |
| 128 | NA | chr1 | + | 825138 | 849592 | ENST00000624927 | 3 | 677 | ENSG00000228794 | LINC01128 | NA | NA | NA | NA | long intergenic non-protein coding RNA 1128 [Source:HGNC Symbol;Acc:HGNC:49377] | 0.96 | 0.12 | 0.89 | 0.11 |
| 129 | NA | chr1 | - | 826206 | 827522 | ENST00000473798 | 1 | 1317 | ENSG00000225880 | LINC00115 | NA | NA | NA | NA | long intergenic non-protein coding RNA 115 [Source:HGNC Symbol;Acc:HGNC:26211] | 2.61 | 0.31 | 0 | 0 |
| 131 | NA | chr1 | + | 827608 | 859446 | ENST00000445118 | 5 | 6606 | ENSG00000228794 | LINC01128 | NA | NA | NA | NA | long intergenic non-protein coding RNA 1128 [Source:HGNC Symbol;Acc:HGNC:49377] | 3.73 | 0.45 | 3.45 | 0.44 |
| 140 | NA | chr1 | + | 827721 | 852090 | ENST00000609009 | 6 | 767 | ENSG00000228794 | LINC01128 | NA | NA | NA | NA | long intergenic non-protein coding RNA 1128 [Source:HGNC Symbol;Acc:HGNC:49377] | 0.65 | 0.08 | 1.22 | 0.16 |
| 142 | NA | chr1 | + | 831605 | 842020 | ENST00000415295 | 2 | 894 | ENSG00000228794 | LINC01128 | NA | NA | NA | NA | long intergenic non-protein coding RNA 1128 [Source:HGNC Symbol;Acc:HGNC:49377] | 3.48 | 0.42 | 4.98 | 0.64 |
| 144 | NA | chr1 | - | 868071 | 876903 | ENST00000446136 | 3 | 1807 | ENSG00000230368 | FAM41C | NA | NA | NA | NA | family with sequence similarity 41 member C [Source:HGNC Symbol;Acc:HGNC:27635] | 0.06 | 0.01 | 0 | 0 |
| 147 | NA | chr1 | + | 873292 | 874349 | ENST00000415481 | 1 | 1058 | ENSG00000234711 | TUBB8P11 | NA | NA | NA | NA | tubulin beta 8 class VIII pseudogene 11 [Source:HGNC Symbol;Acc:HGNC:32337] | 13.50378 | 1.63 | 10.740076 | 1.38 |
| 148 | NA | chr1 | - | 874529 | 877234 | ENST00000635557 | 5 | 837 | ENSG00000283040 | AL669831.6 | NA | NA | NA | NA | NA | 0 | 0 | 0.36 | 0.05 |
| 149 | NA | chr1 | + | 904834 | 915976 | ENST00000607769 | 2 | 351 | ENSG00000272438 | AL645608.7 | NA | NA | NA | NA | NA | 0 | 0 | 0.32 | 0.04 |
| 150 | NA | chr1 | + | 911435 | 914948 | ENST00000448179 | 2 | 3043 | ENSG00000230699 | AL645608.3 | NA | NA | NA | NA | NA | 0.39 | 0.05 | 0.49 | 0.06 |
| 152 | NA | chr1 | - | 916865 | 921016 | ENST00000609207 | 1 | 4152 | ENSG00000223764 | AL645608.1 | NA | NA | NA | NA | NA | 2.35 | 0.28 | 0 | 0 |
| 155 | NA | chr1 | + | 923928 | 939291 | ENST00000420190 | 7 | 1578 | ENSG00000187634 | SAMD11 | GO:0003677(DNA binding);GO:0005634(nucleus) | NA | NA | NA | sterile alpha motif domain containing 11 [Source:HGNC Symbol;Acc:HGNC:28706] | 9.95 | 1.20 | 5.79 | 0.74 |
| 156 | NA | chr1 | + | 925150 | 935793 | ENST00000437963 | 5 | 387 | ENSG00000187634 | SAMD11 | GO:0003677(DNA binding);GO:0005634(nucleus) | NA | NA | NA | sterile alpha motif domain containing 11 [Source:HGNC Symbol;Acc:HGNC:28706] | 0 | 0 | 0.43 | 0.05 |
| 165 | NA | chr1 | + | 925741 | 944581 | ENST00000622503 | 14 | 2557 | ENSG00000187634 | SAMD11 | GO:0003677(DNA binding);GO:0005634(nucleus) | NA | NA | NA | sterile alpha motif domain containing 11 [Source:HGNC Symbol;Acc:HGNC:28706] | 3.30 | 0.40 | 3.25 | 0.42 |
| 169 | NA | chr1 | + | 941076 | 942994 | ENST00000474461 | 4 | 862 | ENSG00000187634 | SAMD11 | GO:0003677(DNA binding);GO:0005634(nucleus) | NA | NA | NA | sterile alpha motif domain containing 11 [Source:HGNC Symbol;Acc:HGNC:28706] | 0.79 | 0.10 | 8.07 | 1.04 |
| 173 | NA | chr1 | - | 944204 | 959290 | ENST00000327044 | 19 | 2790 | ENSG00000188976 | NOC2L | GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005730(nucleolus);GO:0005829(cytosol);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0005515(protein binding);GO:0006915(apoptotic process);GO:0005654(nucleoplasm);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0003714(transcription corepressor activity);GO:0003682(chromatin binding);GO:0042273(ribosomal large subunit biogenesis);GO:0042393(histone binding);GO:1901796(regulation of signal transduction by p53 class mediator);GO:0070491(repressing transcription factor binding);GO:2001243(negative regulation of intrinsic apoptotic signaling pathway);GO:0034644(cellular response to UV);GO:0031497(chromatin assembly);GO:0035067(negative regulation of histone acetylation);GO:0002903(negative regulation of B cell apoptotic process);GO:0031491(nucleosome binding);GO:0030690(Noc1p-Noc2p complex);GO:0030691(Noc2p-Noc3p complex) | NA | NA | NA | NOC2 like nucleolar associated transcriptional repressor [Source:HGNC Symbol;Acc:HGNC:24517] | 42.149437 | 5.08 | 5.28 | 0.68 |
| 174 | NA | chr1 | - | 944205 | 958458 | ENST00000477976 | 17 | 4201 | ENSG00000188976 | NOC2L | GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005730(nucleolus);GO:0005829(cytosol);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0005515(protein binding);GO:0006915(apoptotic process);GO:0005654(nucleoplasm);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0003714(transcription corepressor activity);GO:0003682(chromatin binding);GO:0042273(ribosomal large subunit biogenesis);GO:0042393(histone binding);GO:1901796(regulation of signal transduction by p53 class mediator);GO:0070491(repressing transcription factor binding);GO:2001243(negative regulation of intrinsic apoptotic signaling pathway);GO:0034644(cellular response to UV);GO:0031497(chromatin assembly);GO:0035067(negative regulation of histone acetylation);GO:0002903(negative regulation of B cell apoptotic process);GO:0031491(nucleosome binding);GO:0030690(Noc1p-Noc2p complex);GO:0030691(Noc2p-Noc3p complex) | NA | NA | NA | NOC2 like nucleolar associated transcriptional repressor [Source:HGNC Symbol;Acc:HGNC:24517] | 0.26 | 0.03 | 24.165533 | 3.11 |
| 175 | NA | chr1 | - | 945319 | 945562 | ENST00000496938 | 2 | 149 | ENSG00000188976 | NOC2L | GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005730(nucleolus);GO:0005829(cytosol);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0005515(protein binding);GO:0006915(apoptotic process);GO:0005654(nucleoplasm);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0003714(transcription corepressor activity);GO:0003682(chromatin binding);GO:0042273(ribosomal large subunit biogenesis);GO:0042393(histone binding);GO:1901796(regulation of signal transduction by p53 class mediator);GO:0070491(repressing transcription factor binding);GO:2001243(negative regulation of intrinsic apoptotic signaling pathway);GO:0034644(cellular response to UV);GO:0031497(chromatin assembly);GO:0035067(negative regulation of histone acetylation);GO:0002903(negative regulation of B cell apoptotic process);GO:0031491(nucleosome binding);GO:0030690(Noc1p-Noc2p complex);GO:0030691(Noc2p-Noc3p complex) | NA | NA | NA | NOC2 like nucleolar associated transcriptional repressor [Source:HGNC Symbol;Acc:HGNC:24517] | 0 | 0 | 0.69 | 0.09 |
| 177 | NA | chr1 | - | 958246 | 959256 | ENST00000469563 | 2 | 878 | ENSG00000188976 | NOC2L | GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005730(nucleolus);GO:0005829(cytosol);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0005515(protein binding);GO:0006915(apoptotic process);GO:0005654(nucleoplasm);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0003714(transcription corepressor activity);GO:0003682(chromatin binding);GO:0042273(ribosomal large subunit biogenesis);GO:0042393(histone binding);GO:1901796(regulation of signal transduction by p53 class mediator);GO:0070491(repressing transcription factor binding);GO:2001243(negative regulation of intrinsic apoptotic signaling pathway);GO:0034644(cellular response to UV);GO:0031497(chromatin assembly);GO:0035067(negative regulation of histone acetylation);GO:0002903(negative regulation of B cell apoptotic process);GO:0031491(nucleosome binding);GO:0030690(Noc1p-Noc2p complex);GO:0030691(Noc2p-Noc3p complex) | NA | NA | NA | NOC2 like nucleolar associated transcriptional repressor [Source:HGNC Symbol;Acc:HGNC:24517] | 0.78 | 0.09 | 1.03 | 0.13 |
| 178 | NA | chr1 | + | 960587 | 965715 | ENST00000338591 | 12 | 2560 | ENSG00000187961 | KLHL17 | GO:0016020(membrane);GO:0016567(protein ubiquitination);GO:0003779(actin binding);GO:0005886(plasma membrane);GO:0005615(extracellular space);GO:0030054(cell junction);GO:0045202(synapse);GO:0045211(postsynaptic membrane);GO:0004842(ubiquitin-protein transferase activity);GO:0051015(actin filament binding);GO:0031463(Cul3-RING ubiquitin ligase complex);GO:0014069(postsynaptic density);GO:0015629(actin cytoskeleton);GO:0030036(actin cytoskeleton organization);GO:0007420(brain development);GO:0032947(protein complex scaffold);GO:0032839(dendrite cytoplasm);GO:0031208(POZ domain binding);GO:0043025(neuronal cell body) | NA | NA | NA | kelch like family member 17 [Source:HGNC Symbol;Acc:HGNC:24023] | 15.562367 | 1.88 | 19.251999 | 2.48 |
| 183 | NA | chr1 | + | 966497 | 975108 | ENST00000379410 | 16 | 2404 | ENSG00000187583 | PLEKHN1 | GO:0016020(membrane);GO:0005886(plasma membrane);GO:0005515(protein binding) | NA | NA | NA | pleckstrin homology domain containing N1 [Source:HGNC Symbol;Acc:HGNC:25284] | 7.74 | 0.93 | 11.267826 | 1.45 |
| 186 | NA | chr1 | + | 970875 | 971523 | ENST00000480267 | 3 | 464 | ENSG00000187583 | PLEKHN1 | GO:0016020(membrane);GO:0005886(plasma membrane);GO:0005515(protein binding) | NA | NA | NA | pleckstrin homology domain containing N1 [Source:HGNC Symbol;Acc:HGNC:25284] | 0.15 | 0.02 | 0 | 0 |
| 188 | NA | chr1 | - | 975204 | 982093 | ENST00000341290 | 5 | 3035 | ENSG00000187642 | PERM1 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0014850(response to muscle activity) | NA | NA | NA | PPARGC1 and ESRR induced regulator, muscle 1 [Source:HGNC Symbol;Acc:HGNC:28208] | 4.04 | 0.49 | 2.53 | 0.32 |
| 191 | NA | chr1 | - | 995966 | 998051 | ENST00000606034 | 1 | 2086 | ENSG00000272512 | AL645608.8 | NA | NA | NA | NA | NA | 1.29 | 0.16 | 0 | 0 |
| 192 | NA | chr1 | - | 998962 | 1000172 | ENST00000428771 | 3 | 1040 | ENSG00000188290 | HES4 | GO:0005634(nucleus);GO:0046983(protein dimerization activity);GO:0006355(regulation of transcription, DNA-templated);GO:0006351(transcription, DNA-templated);GO:0003677(DNA binding);GO:0007275(multicellular organism development);GO:0007399(nervous system development);GO:0030154(cell differentiation);GO:0008134(transcription factor binding) | 05165(Human papillomavirus infection) | K09089 | NA | hes family bHLH transcription factor 4 [Source:HGNC Symbol;Acc:HGNC:24149] | 20.661663 | 2.49 | 16.799143 | 2.16 |
| 196 | NA | chr1 | + | 1001138 | 1014541 | ENST00000624697 | 3 | 788 | ENSG00000187608 | ISG15 | GO:0005737(cytoplasm);GO:0005515(protein binding);GO:0005576(extracellular region);GO:0002376(immune system process);GO:0045087(innate immune response);GO:0042742(defense response to bacterium);GO:0051607(defense response to virus);GO:0005654(nucleoplasm);GO:0005829(cytosol);GO:0045071(negative regulation of viral genome replication);GO:0016032(viral process);GO:0030501(positive regulation of bone mineralization);GO:0031397(negative regulation of protein ubiquitination);GO:0060337(type I interferon signaling pathway);GO:0019985(translesion synthesis);GO:0032480(negative regulation of type I interferon production);GO:0031386(protein tag);GO:0019941(modification-dependent protein catabolic process);GO:0032020(ISG15-protein conjugation);GO:0032649(regulation of interferon-gamma production);GO:0034340(response to type I interferon);GO:0045648(positive regulation of erythrocyte differentiation) | 04622(RIG-I-like receptor signaling pathway);05165(Human papillomavirus infection) | NA | NA | ISG15 ubiquitin-like modifier [Source:HGNC Symbol;Acc:HGNC:4053] | 25.238611 | 3.04 | 5.76 | 0.74 |
| 199 | NA | chr1 | - | 1011997 | 1013193 | ENST00000458555 | 2 | 471 | ENSG00000224969 | AL645608.2 | NA | NA | NA | NA | NA | 0 | 0 | 1.22 | 0.16 |
| 200 | NA | chr1 | + | 1013423 | 1014540 | ENST00000379389 | 2 | 711 | ENSG00000187608 | ISG15 | GO:0005737(cytoplasm);GO:0005515(protein binding);GO:0005576(extracellular region);GO:0002376(immune system process);GO:0045087(innate immune response);GO:0042742(defense response to bacterium);GO:0051607(defense response to virus);GO:0005654(nucleoplasm);GO:0005829(cytosol);GO:0045071(negative regulation of viral genome replication);GO:0016032(viral process);GO:0030501(positive regulation of bone mineralization);GO:0031397(negative regulation of protein ubiquitination);GO:0060337(type I interferon signaling pathway);GO:0019985(translesion synthesis);GO:0032480(negative regulation of type I interferon production);GO:0031386(protein tag);GO:0019941(modification-dependent protein catabolic process);GO:0032020(ISG15-protein conjugation);GO:0032649(regulation of interferon-gamma production);GO:0034340(response to type I interferon);GO:0045648(positive regulation of erythrocyte differentiation) | 04622(RIG-I-like receptor signaling pathway);05165(Human papillomavirus infection) | K12159 | NA | ISG15 ubiquitin-like modifier [Source:HGNC Symbol;Acc:HGNC:4053] | 16.181751 | 1.95 | 28.473471 | 3.66 |
| 201 | NA | chr1 | + | 1020123 | 1056116 | ENST00000379370 | 36 | 7323 | ENSG00000188157 | AGRN | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005829(cytosol);GO:0005509(calcium ion binding);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0007010(cytoskeleton organization);GO:0007165(signal transduction);GO:0005576(extracellular region);GO:0043236(laminin binding);GO:0007275(multicellular organism development);GO:0005578(proteinaceous extracellular matrix);GO:0030054(cell junction);GO:0045202(synapse);GO:0030154(cell differentiation);GO:0005200(structural constituent of cytoskeleton);GO:0007213(G-protein coupled acetylcholine receptor signaling pathway);GO:0043113(receptor clustering);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0070062(extracellular exosome);GO:0043547(positive regulation of GTPase activity);GO:0031012(extracellular matrix);GO:0043202(lysosomal lumen);GO:0002162(dystroglycan binding);GO:0033691(sialic acid binding);GO:0035374(chondroitin sulfate binding);GO:0043395(heparan sulfate proteoglycan binding);GO:0001523(retinoid metabolic process);GO:0006024(glycosaminoglycan biosynthetic process);GO:0006027(glycosaminoglycan catabolic process);GO:0030198(extracellular matrix organization);GO:0030203(glycosaminoglycan metabolic process);GO:0045162(clustering of voltage-gated sodium channels);GO:0045887(positive regulation of synaptic growth at neuromuscular junction);GO:0050808(synapse organization);GO:0051491(positive regulation of filopodium assembly);GO:0005605(basal lamina);GO:0005796(Golgi lumen) | 04512(ECM-receptor interaction) | K06254 | NA | agrin [Source:HGNC Symbol;Acc:HGNC:329] | 14.286993 | 1.72 | 31.335733 | 4.03 |
| 202 | NA | chr1 | + | 1020123 | 1056118 | ENST00000620552 | 39 | 7394 | ENSG00000188157 | AGRN | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005829(cytosol);GO:0005509(calcium ion binding);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0007010(cytoskeleton organization);GO:0007165(signal transduction);GO:0005576(extracellular region);GO:0043236(laminin binding);GO:0007275(multicellular organism development);GO:0005578(proteinaceous extracellular matrix);GO:0030054(cell junction);GO:0045202(synapse);GO:0030154(cell differentiation);GO:0005200(structural constituent of cytoskeleton);GO:0007213(G-protein coupled acetylcholine receptor signaling pathway);GO:0043113(receptor clustering);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0070062(extracellular exosome);GO:0043547(positive regulation of GTPase activity);GO:0031012(extracellular matrix);GO:0043202(lysosomal lumen);GO:0002162(dystroglycan binding);GO:0033691(sialic acid binding);GO:0035374(chondroitin sulfate binding);GO:0043395(heparan sulfate proteoglycan binding);GO:0001523(retinoid metabolic process);GO:0006024(glycosaminoglycan biosynthetic process);GO:0006027(glycosaminoglycan catabolic process);GO:0030198(extracellular matrix organization);GO:0030203(glycosaminoglycan metabolic process);GO:0045162(clustering of voltage-gated sodium channels);GO:0045887(positive regulation of synaptic growth at neuromuscular junction);GO:0050808(synapse organization);GO:0051491(positive regulation of filopodium assembly);GO:0005605(basal lamina);GO:0005796(Golgi lumen) | 04512(ECM-receptor interaction) | K06254 | NA | agrin [Source:HGNC Symbol;Acc:HGNC:329] | 313.871216 | 37.84407 | 251.318115 | 32.333313 |
| 203 | NA | chr1 | + | 1034106 | 1040725 | ENST00000477585 | 3 | 707 | ENSG00000188157 | AGRN | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005829(cytosol);GO:0005509(calcium ion binding);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0007010(cytoskeleton organization);GO:0007165(signal transduction);GO:0005576(extracellular region);GO:0043236(laminin binding);GO:0007275(multicellular organism development);GO:0005578(proteinaceous extracellular matrix);GO:0030054(cell junction);GO:0045202(synapse);GO:0030154(cell differentiation);GO:0005200(structural constituent of cytoskeleton);GO:0007213(G-protein coupled acetylcholine receptor signaling pathway);GO:0043113(receptor clustering);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0070062(extracellular exosome);GO:0043547(positive regulation of GTPase activity);GO:0031012(extracellular matrix);GO:0043202(lysosomal lumen);GO:0002162(dystroglycan binding);GO:0033691(sialic acid binding);GO:0035374(chondroitin sulfate binding);GO:0043395(heparan sulfate proteoglycan binding);GO:0001523(retinoid metabolic process);GO:0006024(glycosaminoglycan biosynthetic process);GO:0006027(glycosaminoglycan catabolic process);GO:0030198(extracellular matrix organization);GO:0030203(glycosaminoglycan metabolic process);GO:0045162(clustering of voltage-gated sodium channels);GO:0045887(positive regulation of synaptic growth at neuromuscular junction);GO:0050808(synapse organization);GO:0051491(positive regulation of filopodium assembly);GO:0005605(basal lamina);GO:0005796(Golgi lumen) | 04512(ECM-receptor interaction) | NA | NA | agrin [Source:HGNC Symbol;Acc:HGNC:329] | 2.62 | 0.32 | 0.82 | 0.11 |
| 205 | NA | chr1 | + | 1045399 | 1046349 | ENST00000479707 | 4 | 584 | ENSG00000188157 | AGRN | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005829(cytosol);GO:0005509(calcium ion binding);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0007010(cytoskeleton organization);GO:0007165(signal transduction);GO:0005576(extracellular region);GO:0043236(laminin binding);GO:0007275(multicellular organism development);GO:0005578(proteinaceous extracellular matrix);GO:0030054(cell junction);GO:0045202(synapse);GO:0030154(cell differentiation);GO:0005200(structural constituent of cytoskeleton);GO:0007213(G-protein coupled acetylcholine receptor signaling pathway);GO:0043113(receptor clustering);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0070062(extracellular exosome);GO:0043547(positive regulation of GTPase activity);GO:0031012(extracellular matrix);GO:0043202(lysosomal lumen);GO:0002162(dystroglycan binding);GO:0033691(sialic acid binding);GO:0035374(chondroitin sulfate binding);GO:0043395(heparan sulfate proteoglycan binding);GO:0001523(retinoid metabolic process);GO:0006024(glycosaminoglycan biosynthetic process);GO:0006027(glycosaminoglycan catabolic process);GO:0030198(extracellular matrix organization);GO:0030203(glycosaminoglycan metabolic process);GO:0045162(clustering of voltage-gated sodium channels);GO:0045887(positive regulation of synaptic growth at neuromuscular junction);GO:0050808(synapse organization);GO:0051491(positive regulation of filopodium assembly);GO:0005605(basal lamina);GO:0005796(Golgi lumen) | 04512(ECM-receptor interaction) | NA | NA | agrin [Source:HGNC Symbol;Acc:HGNC:329] | 4.92 | 0.59 | 2.74 | 0.35 |
| 208 | NA | chr1 | + | 1048529 | 1049394 | ENST00000492947 | 2 | 690 | ENSG00000188157 | AGRN | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005829(cytosol);GO:0005509(calcium ion binding);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0007010(cytoskeleton organization);GO:0007165(signal transduction);GO:0005576(extracellular region);GO:0043236(laminin binding);GO:0007275(multicellular organism development);GO:0005578(proteinaceous extracellular matrix);GO:0030054(cell junction);GO:0045202(synapse);GO:0030154(cell differentiation);GO:0005200(structural constituent of cytoskeleton);GO:0007213(G-protein coupled acetylcholine receptor signaling pathway);GO:0043113(receptor clustering);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0070062(extracellular exosome);GO:0043547(positive regulation of GTPase activity);GO:0031012(extracellular matrix);GO:0043202(lysosomal lumen);GO:0002162(dystroglycan binding);GO:0033691(sialic acid binding);GO:0035374(chondroitin sulfate binding);GO:0043395(heparan sulfate proteoglycan binding);GO:0001523(retinoid metabolic process);GO:0006024(glycosaminoglycan biosynthetic process);GO:0006027(glycosaminoglycan catabolic process);GO:0030198(extracellular matrix organization);GO:0030203(glycosaminoglycan metabolic process);GO:0045162(clustering of voltage-gated sodium channels);GO:0045887(positive regulation of synaptic growth at neuromuscular junction);GO:0050808(synapse organization);GO:0051491(positive regulation of filopodium assembly);GO:0005605(basal lamina);GO:0005796(Golgi lumen) | 04512(ECM-receptor interaction) | NA | NA | agrin [Source:HGNC Symbol;Acc:HGNC:329] | 3.30 | 0.40 | 1.77 | 0.23 |
| 210 | NA | chr1 | + | 1051986 | 1056112 | ENST00000461111 | 3 | 3385 | ENSG00000188157 | AGRN | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005829(cytosol);GO:0005509(calcium ion binding);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0007010(cytoskeleton organization);GO:0007165(signal transduction);GO:0005576(extracellular region);GO:0043236(laminin binding);GO:0007275(multicellular organism development);GO:0005578(proteinaceous extracellular matrix);GO:0030054(cell junction);GO:0045202(synapse);GO:0030154(cell differentiation);GO:0005200(structural constituent of cytoskeleton);GO:0007213(G-protein coupled acetylcholine receptor signaling pathway);GO:0043113(receptor clustering);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0070062(extracellular exosome);GO:0043547(positive regulation of GTPase activity);GO:0031012(extracellular matrix);GO:0043202(lysosomal lumen);GO:0002162(dystroglycan binding);GO:0033691(sialic acid binding);GO:0035374(chondroitin sulfate binding);GO:0043395(heparan sulfate proteoglycan binding);GO:0001523(retinoid metabolic process);GO:0006024(glycosaminoglycan biosynthetic process);GO:0006027(glycosaminoglycan catabolic process);GO:0030198(extracellular matrix organization);GO:0030203(glycosaminoglycan metabolic process);GO:0045162(clustering of voltage-gated sodium channels);GO:0045887(positive regulation of synaptic growth at neuromuscular junction);GO:0050808(synapse organization);GO:0051491(positive regulation of filopodium assembly);GO:0005605(basal lamina);GO:0005796(Golgi lumen) | 04512(ECM-receptor interaction) | NA | NA | agrin [Source:HGNC Symbol;Acc:HGNC:329] | 6.41 | 0.77 | 0 | 0 |
| 212 | NA | chr1 | + | 1059734 | 1066456 | ENST00000427998 | 4 | 977 | ENSG00000217801 | AL390719.1 | NA | NA | NA | NA | NA | 25.348156 | 3.06 | 22.769318 | 2.93 |
| 213 | NA | chr1 | + | 1061020 | 1066005 | ENST00000394517 | 3 | 397 | ENSG00000217801 | AL390719.1 | NA | NA | NA | NA | NA | 5.15 | 0.62 | 4.00 | 0.51 |
| 214 | NA | chr1 | - | 1062208 | 1063288 | ENST00000442292 | 2 | 829 | ENSG00000273443 | AL645608.9 | NA | NA | NA | NA | NA | 8.39 | 1.01 | 5.20 | 0.67 |
| 216 | NA | chr1 | + | 1063079 | 1069355 | ENST00000412397 | 10 | 1219 | ENSG00000217801 | AL390719.1 | NA | NA | NA | NA | NA | 1.37 | 0.16 | 2.14 | 0.28 |
| 219 | NA | chr1 | - | 1070966 | 1074307 | ENST00000453464 | 2 | 1902 | ENSG00000237330 | RNF223 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0046872(metal ion binding) | NA | NA | NA | ring finger protein 223 [Source:HGNC Symbol;Acc:HGNC:40020] | 0 | 0 | 0.25 | 0.03 |
| 220 | NA | chr1 | - | 1081818 | 1116081 | ENST00000379339 | 12 | 2429 | ENSG00000131591 | C1orf159 | GO:0016020(membrane);GO:0016021(integral component of membrane) | NA | NA | NA | chromosome 1 open reading frame 159 [Source:HGNC Symbol;Acc:HGNC:26062] | 3.99 | 0.48 | 0.13 | 0.02 |
| 224 | NA | chr1 | - | 1081825 | 1087636 | ENST00000487177 | 3 | 3315 | ENSG00000131591 | C1orf159 | GO:0016020(membrane);GO:0016021(integral component of membrane) | NA | NA | NA | chromosome 1 open reading frame 159 [Source:HGNC Symbol;Acc:HGNC:26062] | 39.874851 | 4.81 | 39.195755 | 5.04 |
| 226 | NA | chr1 | - | 1081825 | 1116089 | ENST00000477196 | 10 | 3214 | ENSG00000131591 | C1orf159 | GO:0016020(membrane);GO:0016021(integral component of membrane) | NA | NA | NA | chromosome 1 open reading frame 159 [Source:HGNC Symbol;Acc:HGNC:26062] | 0 | 0 | 1.49 | 0.19 |
| 228 | NA | chr1 | - | 1082235 | 1092174 | ENST00000379320 | 8 | 2324 | ENSG00000131591 | C1orf159 | GO:0016020(membrane);GO:0016021(integral component of membrane) | NA | NA | NA | chromosome 1 open reading frame 159 [Source:HGNC Symbol;Acc:HGNC:26062] | 2.75 | 0.33 | 4.29 | 0.55 |
| 229 | NA | chr1 | - | 1082700 | 1106127 | ENST00000379319 | 10 | 1097 | ENSG00000131591 | C1orf159 | GO:0016020(membrane);GO:0016021(integral component of membrane) | NA | NA | NA | chromosome 1 open reading frame 159 [Source:HGNC Symbol;Acc:HGNC:26062] | 0.26 | 0.03 | 0.74 | 0.09 |
| 233 | NA | chr1 | - | 1087141 | 1092294 | ENST00000473600 | 5 | 793 | ENSG00000131591 | C1orf159 | GO:0016020(membrane);GO:0016021(integral component of membrane) | NA | NA | NA | chromosome 1 open reading frame 159 [Source:HGNC Symbol;Acc:HGNC:26062] | 0 | 0 | 1.02 | 0.13 |
| 236 | NA | chr1 | - | 1090406 | 1092813 | ENST00000427787 | 5 | 575 | ENSG00000131591 | C1orf159 | GO:0016020(membrane);GO:0016021(integral component of membrane) | NA | NA | NA | chromosome 1 open reading frame 159 [Source:HGNC Symbol;Acc:HGNC:26062] | 1.50 | 0.18 | 0 | 0 |
| 244 | NA | chr1 | - | 1173056 | 1179555 | ENST00000379317 | 2 | 3532 | ENSG00000205231 | TTLL10-AS1 | NA | NA | NA | NA | TTLL10 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:41159] | 0 | 0 | 0.13 | 0.02 |
| 246 | NA | chr1 | + | 1173903 | 1197935 | ENST00000379290 | 16 | 2283 | ENSG00000162571 | TTLL10 | GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0006464(cellular protein modification process);GO:0005515(protein binding);GO:0005829(cytosol);GO:0070735(protein-glycine ligase activity);GO:0018094(protein polyglycylation) | NA | NA | NA | tubulin tyrosine ligase like 10 [Source:HGNC Symbol;Acc:HGNC:26693] | 0.47 | 0.06 | 0.12 | 0.02 |
| 248 | NA | chr1 | + | 1179697 | 1185864 | ENST00000379288 | 9 | 2114 | ENSG00000162571 | TTLL10 | GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0006464(cellular protein modification process);GO:0005515(protein binding);GO:0005829(cytosol);GO:0070735(protein-glycine ligase activity);GO:0018094(protein polyglycylation) | NA | NA | NA | tubulin tyrosine ligase like 10 [Source:HGNC Symbol;Acc:HGNC:26693] | 0.14 | 0.02 | 0 | 0 |
| 261 | NA | chr1 | - | 1216931 | 1232031 | ENST00000465727 | 7 | 2095 | ENSG00000078808 | SDF4 | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0005794(Golgi apparatus);GO:0046872(metal ion binding);GO:0005509(calcium ion binding);GO:0045444(fat cell differentiation);GO:0005770(late endosome);GO:0005796(Golgi lumen);GO:0005886(plasma membrane);GO:0042995(cell projection);GO:0006887(exocytosis);GO:0070062(extracellular exosome);GO:0042802(identical protein binding);GO:0032059(bleb);GO:0045471(response to ethanol);GO:0009650(UV protection);GO:0021549(cerebellum development);GO:0017156(calcium ion regulated exocytosis);GO:0070625(zymogen granule exocytosis) | NA | NA | NA | stromal cell derived factor 4 [Source:HGNC Symbol;Acc:HGNC:24188] | 348.803741 | 42.055954 | 309.044067 | 39.76004 |
| 263 | NA | chr1 | - | 1216934 | 1220293 | ENST00000478938 | 2 | 2591 | ENSG00000078808 | SDF4 | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0005794(Golgi apparatus);GO:0046872(metal ion binding);GO:0005509(calcium ion binding);GO:0045444(fat cell differentiation);GO:0005770(late endosome);GO:0005796(Golgi lumen);GO:0005886(plasma membrane);GO:0042995(cell projection);GO:0006887(exocytosis);GO:0070062(extracellular exosome);GO:0042802(identical protein binding);GO:0032059(bleb);GO:0045471(response to ethanol);GO:0009650(UV protection);GO:0021549(cerebellum development);GO:0017156(calcium ion regulated exocytosis);GO:0070625(zymogen granule exocytosis) | NA | NA | NA | stromal cell derived factor 4 [Source:HGNC Symbol;Acc:HGNC:24188] | 1.21 | 0.15 | 1.35 | 0.17 |
| 266 | NA | chr1 | + | 1232265 | 1235041 | ENST00000379198 | 1 | 2777 | ENSG00000176022 | B3GALT6 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0016740(transferase activity);GO:0016757(transferase activity, transferring glycosyl groups);GO:0006486(protein glycosylation);GO:0005794(Golgi apparatus);GO:0008378(galactosyltransferase activity);GO:0032580(Golgi cisterna membrane);GO:0000139(Golgi membrane);GO:0030203(glycosaminoglycan metabolic process);GO:0015012(heparan sulfate proteoglycan biosynthetic process);GO:0006024(glycosaminoglycan biosynthetic process);GO:0005797(Golgi medial cisterna);GO:0035250(UDP-galactosyltransferase activity);GO:0030206(chondroitin sulfate biosynthetic process);GO:0008499(UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity);GO:0047220(galactosylxylosylprotein 3-beta-galactosyltransferase activity) | 00532(Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate);00534(Glycosaminoglycan biosynthesis - heparan sulfate / heparin) | K00734 | EC:2.4.1.134 | beta-1,3-galactosyltransferase 6 [Source:HGNC Symbol;Acc:HGNC:17978] | 0 | 0 | 67.904305 | 8.74 |
| 269 | NA | chr1 | - | 1242453 | 1246722 | ENST00000330388 | 8 | 1036 | ENSG00000184163 | C1QTNF12 | GO:0005576(extracellular region);GO:0005179(hormone activity);GO:0010906(regulation of glucose metabolic process);GO:0035774(positive regulation of insulin secretion involved in cellular response to glucose stimulus);GO:0045721(negative regulation of gluconeogenesis);GO:0046324(regulation of glucose import);GO:0046326(positive regulation of glucose import);GO:0046628(positive regulation of insulin receptor signaling pathway);GO:0050728(negative regulation of inflammatory response);GO:0051897(positive regulation of protein kinase B signaling);GO:0005615(extracellular space) | NA | NA | NA | C1q and TNF related 12 [Source:HGNC Symbol;Acc:HGNC:32308] | 0.91 | 0.11 | 0.67 | 0.09 |
Definition of table:
| term | Description |
|---|---|
| chr | The name of the chromosome (e.g. chr3, chrY, chr2 random) or scaffold (e.g. scaffold10671) |
| strand | sense strand="+"; antisense strand="-" |
| start | The starting position of transcript in the chromosome or scaffold |
| end | The ending position of transcript in the chromosome or scaffold |
| t_name | The name of transcript |
| num_exons | number of exons |
| length | length of transcript |
| gene_id | The ID of gene, some ids started with "MSTRG" are named by transcript assembly software StringTie |
| gene_name | The name of gene,or gene symbol |
| GO | Gene ontology term and description http://www.geneontology.org/ |
| KEGG | KEGG pathway annotation and description http://www.kegg.jp/kegg/kegg3a.html |
| KO_Entry | KO functional ortholog, KO groups identified by K numbers and, in metabolic maps |
| EC | enzyme |
| Description | Description of gene |
| cov.A | count number or coverage of transcript |
| FPKM.A | Normalization of gene expression level by FPKM value |
Definition of class code:
| Class code | Description |
|---|---|
| = | Complete match (mRNA, lncRNA etc) |
| c | Contained |
| j | Potentially novel isoform (fragment): at least one splice junction is hared with a reference transcript |
| e | Single exon transfrag overlapping a reference exon and at least 10 bp of a reference intron, indicating a possible pre-mRNA fragment. |
| i | A transfrag falling entirely within a reference intron |
| o | Generic exonic overlap with a reference transcript |
| p | Possible polymerase run-on fragment (within 2Kbases of a reference transcript) |
| r | Repeat. Currently determined by looking at the soft-masked reference sequence and applied to transcripts where at least 50% of the are lower casebases |
| u | Unknown, intergenic transcript |
| x | Exonic overlap with reference on the opposite strand |
| s | An intron of the transfrag overlaps a reference intron on the opposite strand (likely due to read mapping errors) |
| . | (.tracking file only, indicates multiple classifications) |
document location: summary6_transcript_expression/5_isoforms_fpkm_expression.xlsx
Density:
Boxplot:
document location
summary6_transcript_expression/*_gene_expression_density.png
summary6_transcript_expression/*_transcript_expression_density.png
summary6_transcript_expression/*_gene_expression_boxplot.png
summary6_transcript_expression/*_transcript_expression_boxplot.png
Exploratory analysis, visualization and statistical modeling are the next steps after assembling and quantifying genes and transcripts from StringTie. The assembly of input reads reconstructs the exon–intron structure of genes and their isoforms. R package edgeR takes both transcript and gene expression levels from StringTie and applies rigorous statistical methods to determine which transcripts and genes are differentially expressed between two or more experiments. edgeR also includes plotting tools as part of the R/Bioconductor package that help visualize the results.
| gene_id | gene_name | transcript_id | GO | KEGG | KO_ENTRY | EC | Description | FPKM.M14_OE | FPKM.M14_NC | fc | log2(fc) | pval | qval | regulation | significant |
| ENSG00000132967 | HMGB1P5 | ENST00000451497 | NA | NA | NA | NA | high mobility group box 1 pseudogene 5 [Source:HGNC Symbol;Acc:HGNC:4997] | 0.83 | 278.68408 | 0.00 | -8.39980990887748 | 2.08e-20 | 2.99e-16 | down | yes |
| ENSG00000166897 | ELFN2 | ENST00000402918;ENST00000451509;ENST00000414347;ENST00000435824;ENST00000424973 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005615(extracellular space);GO:0004864(protein phosphatase inhibitor activity);GO:0019902(phosphatase binding);GO:0010923(negative regulation of phosphatase activity);GO:0010923(negative regulation of phosphatase activity) | NA | NA | NA | extracellular leucine rich repeat and fibronectin type III domain containing 2 [Source:HGNC Symbol;Acc:HGNC:29396] | 0.07 | 11.6113688709861 | 0.01 | -7.40124791982042 | 3.03e-17 | 2.01e-13 | down | yes |
| ENSG00000223361 | FTH1P10 | ENST00000401830 | NA | NA | NA | NA | ferritin heavy chain 1 pseudogene 10 [Source:HGNC Symbol;Acc:HGNC:3980] | 1.64 | 251.200851 | 0.01 | -7.25830254086227 | 4.20e-17 | 2.01e-13 | down | yes |
| ENSG00000145388 | METTL14 | ENST00000388822 | GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005654(nucleoplasm);GO:0008168(methyltransferase activity);GO:0016740(transferase activity);GO:0032259(methylation);GO:0006139(nucleobase-containing compound metabolic process);GO:0003729(mRNA binding);GO:0005515(protein binding);GO:0000398(mRNA splicing, via spliceosome);GO:0016070(RNA metabolic process);GO:0019827(stem cell population maintenance);GO:0061157(mRNA destabilization);GO:0080009(mRNA methylation);GO:0036396(MIS complex);GO:0016422(mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity) | NA | NA | NA | methyltransferase like 14 [Source:HGNC Symbol;Acc:HGNC:29330] | 113.079313256485 | 0.97 | 116.925215255614 | 6.87 | 2.82e-16 | 1.01e-12 | up | yes |
| ENSG00000268173 | AC007192.1 | ENST00000593731 | NA | NA | NA | NA | 0.13 | 15.335408 | 0.01 | -6.83520972451006 | 1.07e-15 | 3.08e-12 | down | yes | |
| ENSG00000180574 | AC068775.1 | ENST00000322446;ENST00000538173 | GO:0005525(GTP binding);GO:0003924(GTPase activity);GO:0003743(translation initiation factor activity);GO:0006412(translation);GO:0006413(translational initiation);GO:0000166(nucleotide binding);GO:0000166(nucleotide binding) | NA | NA | NA | Putative eukaryotic translation initiation factor 2 subunit 3-like protein [Source:UniProtKB/Swiss-Prot;Acc:Q2VIR3] | 0.53 | 56.4813463929014 | 0.01 | -6.7297462449392 | 1.61e-15 | 3.85e-12 | down | yes |
| ENSG00000161654 | LSM12 | ENST00000585388;ENST00000591247 | NA | NA | NA | NA | LSM12 homolog [Source:HGNC Symbol;Acc:HGNC:26407] | 0.24 | 10.9960782646745 | 0.02 | -5.5021336647813 | 9.52e-12 | 1.95e-08 | down | yes |
| ENSG00000240184 | PCDHGC3 | ENST00000622656;ENST00000622836;ENST00000611950 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005509(calcium ion binding);GO:0007155(cell adhesion);GO:0007156(homophilic cell adhesion via plasma membrane adhesion molecules);GO:0005886(plasma membrane);GO:0005887(integral component of plasma membrane);GO:0070062(extracellular exosome);GO:0007399(nervous system development);GO:0007267(cell-cell signaling);GO:0016339(calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules);GO:0016339(calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules) | NA | NA | NA | protocadherin gamma subfamily C, 3 [Source:HGNC Symbol;Acc:HGNC:8716] | 4.02 | 0.10 | 39.5149524376833 | 5.30 | 2.49e-11 | 4.48e-08 | up | yes |
| ENSG00000111328 | CDK2AP1 | ENST00000618072;ENST00000261692;ENST00000538446;ENST00000544658;ENST00000535979 | GO:0005634(nucleus);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0005515(protein binding);GO:0007049(cell cycle);GO:0048471(perinuclear region of cytoplasm);GO:0001934(positive regulation of protein phosphorylation);GO:0070182(DNA polymerase binding);GO:0006261(DNA-dependent DNA replication);GO:0006261(DNA-dependent DNA replication) | NA | NA | NA | cyclin dependent kinase 2 associated protein 1 [Source:HGNC Symbol;Acc:HGNC:14002] | 27.6970164687199 | 0.78 | 35.3062955771881 | 5.14 | 4.02e-11 | 6.43e-08 | up | yes |
| ENSG00000231120 | BTF3P10 | ENST00000433415 | NA | NA | NA | NA | basic transcription factor 3 pseudogene 10 [Source:HGNC Symbol;Acc:HGNC:38570] | 175.663574 | 5.37 | 32.6839666506035 | 5.03 | 7.46e-11 | 1.07e-07 | up | yes |
| ENSG00000167395 | ZNF646 | ENST00000564189;ENST00000394979 | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0005515(protein binding);GO:0005515(protein binding) | NA | NA | NA | zinc finger protein 646 [Source:HGNC Symbol;Acc:HGNC:29004] | 0.12 | 4.07 | 0.03 | -5.11938750288584 | 8.69e-11 | 1.13e-07 | down | yes |
| ENSG00000139146 | FAM60A | ENST00000536836;ENST00000337682;ENST00000454658 | GO:0005634(nucleus);GO:0005515(protein binding);GO:0030336(negative regulation of cell migration);GO:0016580(Sin3 complex);GO:0016580(Sin3 complex) | NA | NA | NA | family with sequence similarity 60 member A [Source:HGNC Symbol;Acc:HGNC:30702] | 0.26 | 8.77 | 0.03 | -5.06808422538973 | 1.08e-10 | 1.29e-07 | down | yes |
| ENSG00000165821 | SALL2 | ENST00000611430;ENST00000614342 | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0005515(protein binding);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0001654(eye development);GO:0007165(signal transduction);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0001228(transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0043565(sequence-specific DNA binding);GO:0000977(RNA polymerase II regulatory region sequence-specific DNA binding);GO:0044212(transcription regulatory region DNA binding);GO:0006366(transcription from RNA polymerase II promoter);GO:0021915(neural tube development);GO:0045892(negative regulation of transcription, DNA-templated);GO:0016581(NuRD complex);GO:0016581(NuRD complex) | NA | NA | NA | spalt like transcription factor 2 [Source:HGNC Symbol;Acc:HGNC:10526] | 3.52 | 0.11 | 32.047147684587 | 5.00 | 1.95e-10 | 2.16e-07 | up | yes |
| ENSG00000131100 | ATP6V1E1 | ENST00000253413;ENST00000460085;ENST00000484653 | GO:0006810(transport);GO:0016787(hydrolase activity);GO:0005765(lysosomal membrane);GO:0005829(cytosol);GO:0006811(ion transport);GO:0015991(ATP hydrolysis coupled proton transport);GO:0015992(proton transport);GO:0046961(proton-transporting ATPase activity, rotational mechanism);GO:0005515(protein binding);GO:0033178(proton-transporting two-sector ATPase complex, catalytic domain);GO:0005768(endosome);GO:0070062(extracellular exosome);GO:0034220(ion transmembrane transport);GO:0016324(apical plasma membrane);GO:0008286(insulin receptor signaling pathway);GO:0016241(regulation of macroautophagy);GO:0033572(transferrin transport);GO:0090383(phagosome acidification);GO:0051117(ATPase binding);GO:0016469(proton-transporting two-sector ATPase complex);GO:0008553(hydrogen-exporting ATPase activity, phosphorylative mechanism);GO:0005737(cytoplasm);GO:0005739(mitochondrion);GO:0005902(microvillus);GO:0005902(microvillus) | 00190(Oxidative phosphorylation);04150(mTOR signaling pathway);04145(Phagosome);04966(Collecting duct acid secretion);04721(Synaptic vesicle cycle);05323(Rheumatoid arthritis);05110(Vibrio cholerae infection);05120(Epithelial cell signaling in Helicobacter pylori infection);05120(Epithelial cell signaling in Helicobacter pylori infection) | K02150 | NA | ATPase H+ transporting V1 subunit E1 [Source:HGNC Symbol;Acc:HGNC:857] | 6.05 | 0.19 | 31.2547387265628 | 4.97 | 2.29e-10 | 2.32e-07 | up | yes |
| ENSG00000147955 | SIGMAR1 | ENST00000497006;ENST00000378892 | GO:0006810(transport);GO:0005634(nucleus);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0005886(plasma membrane);GO:0042995(cell projection);GO:0006869(lipid transport);GO:0030054(cell junction);GO:0045202(synapse);GO:0045211(postsynaptic membrane);GO:0031410(cytoplasmic vesicle);GO:0038003(opioid receptor signaling pathway);GO:0030426(growth cone);GO:0005637(nuclear inner membrane);GO:0014069(postsynaptic density);GO:0005887(integral component of plasma membrane);GO:0005811(lipid particle);GO:0005635(nuclear envelope);GO:0070207(protein homotrimerization);GO:0005640(nuclear outer membrane);GO:0043523(regulation of neuron apoptotic process);GO:0008144(drug binding);GO:0004872(receptor activity);GO:0004985(opioid receptor activity);GO:0007399(nervous system development);GO:0007399(nervous system development) | NA | NA | NA | sigma non-opioid intracellular receptor 1 [Source:HGNC Symbol;Acc:HGNC:8157] | 2.10 | 63.0646359635701 | 0.03 | -4.90761503199783 | 2.42e-10 | 2.32e-07 | down | yes |
| ENSG00000143252 | SDHC | ENST00000367975;ENST00000515731;ENST00000470743 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0046872(metal ion binding);GO:0055114(oxidation-reduction process);GO:0016627(oxidoreductase activity, acting on the CH-CH group of donors);GO:0006099(tricarboxylic acid cycle);GO:0005743(mitochondrial inner membrane);GO:0005739(mitochondrion);GO:0009055(electron carrier activity);GO:0020037(heme binding);GO:0000104(succinate dehydrogenase activity);GO:0045281(succinate dehydrogenase complex);GO:0006121(mitochondrial electron transport, succinate to ubiquinone);GO:0005749(mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone));GO:0009060(aerobic respiration);GO:0045273(respiratory chain complex II);GO:0045273(respiratory chain complex II) | 00020(Citrate cycle (TCA cycle));00190(Oxidative phosphorylation);05010(Alzheimer's disease);05012(Parkinson's disease);05016(Huntington's disease);04932(Non-alcoholic fatty liver disease (NAFLD));04932(Non-alcoholic fatty liver disease (NAFLD)) | K00236 | NA | succinate dehydrogenase complex subunit C [Source:HGNC Symbol;Acc:HGNC:10682] | 0.12 | 3.64 | 0.03 | -4.86835323626821 | 3.69e-10 | 3.31e-07 | down | yes |
| ENSG00000271303 | SRXN1 | ENST00000381962 | GO:0005737(cytoplasm);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0005829(cytosol);GO:0098869(cellular oxidant detoxification);GO:0006979(response to oxidative stress);GO:0034599(cellular response to oxidative stress);GO:0016209(antioxidant activity);GO:0016667(oxidoreductase activity, acting on a sulfur group of donors);GO:0032542(sulfiredoxin activity) | NA | NA | NA | sulfiredoxin 1 [Source:HGNC Symbol;Acc:HGNC:16132] | 62.652645 | 2.45 | 25.5866800509507 | 4.68 | 6.59e-10 | 5.57e-07 | up | yes |
| ENSG00000180764 | PIPSL | ENST00000480546 | NA | NA | NA | NA | PIP5K1A and PSMD4-like, pseudogene [Source:HGNC Symbol;Acc:HGNC:23733] | 54.989426 | 2.18 | 25.2439729901819 | 4.66 | 7.50e-10 | 5.98e-07 | up | yes |
| ENSG00000265241 | RBM8A | ENST00000632040;ENST00000583313 | GO:0005634(nucleus);GO:0003676(nucleic acid binding);GO:0003723(RNA binding);GO:0003729(mRNA binding);GO:0006396(RNA processing);GO:0006397(mRNA processing);GO:0006810(transport);GO:0008380(RNA splicing);GO:0051028(mRNA transport);GO:0005737(cytoplasm);GO:0016607(nuclear speck);GO:0030425(dendrite);GO:0043025(neuronal cell body);GO:0005515(protein binding);GO:0005681(spliceosomal complex);GO:0005654(nucleoplasm);GO:0005829(cytosol);GO:0006417(regulation of translation);GO:0000398(mRNA splicing, via spliceosome);GO:0006369(termination of RNA polymerase II transcription);GO:0006405(RNA export from nucleus);GO:0031124(mRNA 3'-end processing);GO:0035145(exon-exon junction complex);GO:0000381(regulation of alternative mRNA splicing, via spliceosome);GO:0000184(nuclear-transcribed mRNA catabolic process, nonsense-mediated decay);GO:0006406(mRNA export from nucleus);GO:0071013(catalytic step 2 spliceosome);GO:0071013(catalytic step 2 spliceosome) | 03040(Spliceosome);03013(RNA transport);03015(mRNA surveillance pathway);03015(mRNA surveillance pathway) | K12876 | NA | RNA binding motif protein 8A [Source:HGNC Symbol;Acc:HGNC:9905] | 0.20 | 5.24 | 0.04 | -4.68304562231244 | 1.40e-09 | 1.06e-06 | down | yes |
| ENSG00000163807 | KIAA1143 | ENST00000296121;ENST00000484437 | GO:0005515(protein binding);GO:0005515(protein binding) | NA | NA | NA | KIAA1143 [Source:HGNC Symbol;Acc:HGNC:29198] | 0.22 | 5.46 | 0.04 | -4.62878701877958 | 1.95e-09 | 1.40e-06 | down | yes |
| ENSG00000134899 | ERCC5 | ENST00000355739;ENST00000375954;ENST00000375958 | GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0005654(nucleoplasm);GO:0016787(hydrolase activity);GO:0003677(DNA binding);GO:0003824(catalytic activity);GO:0042803(protein homodimerization activity);GO:0004519(endonuclease activity);GO:0006281(DNA repair);GO:0006974(cellular response to DNA damage stimulus);GO:0016788(hydrolase activity, acting on ester bonds);GO:0006289(nucleotide-excision repair);GO:0004518(nuclease activity);GO:0003697(single-stranded DNA binding);GO:0005515(protein binding);GO:0003690(double-stranded DNA binding);GO:0047485(protein N-terminus binding);GO:0043066(negative regulation of apoptotic process);GO:0009411(response to UV);GO:0006283(transcription-coupled nucleotide-excision repair);GO:0005662(DNA replication factor A complex);GO:0016591(DNA-directed RNA polymerase II, holoenzyme);GO:0006293(nucleotide-excision repair, preincision complex stabilization);GO:0006295(nucleotide-excision repair, DNA incision, 3'-to lesion);GO:0006296(nucleotide-excision repair, DNA incision, 5'-to lesion);GO:0033683(nucleotide-excision repair, DNA incision);GO:0006294(nucleotide-excision repair, preincision complex assembly);GO:0009650(UV protection);GO:0005675(holo TFIIH complex);GO:0010225(response to UV-C);GO:0004520(endodeoxyribonuclease activity);GO:0000405(bubble DNA binding);GO:0000405(bubble DNA binding) | 03420(Nucleotide excision repair);03420(Nucleotide excision repair) | K10846 | NA | ERCC excision repair 5, endonuclease [Source:HGNC Symbol;Acc:HGNC:3437] | 0.23 | 5.41 | 0.04 | -4.54149679875712 | 3.23e-09 | 2.21e-06 | down | yes |
| ENSG00000116044 | NFE2L2 | ENST00000397062;ENST00000458603;ENST00000477534;ENST00000423513;ENST00000397063 | GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0005634(nucleus);GO:0071356(cellular response to tumor necrosis factor);GO:2000352(negative regulation of endothelial cell apoptotic process);GO:0005737(cytoplasm);GO:0005794(Golgi apparatus);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0006355(regulation of transcription, DNA-templated);GO:0006351(transcription, DNA-templated);GO:0016567(protein ubiquitination);GO:0003677(DNA binding);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0005886(plasma membrane);GO:0005813(centrosome);GO:0005515(protein binding);GO:0034599(cellular response to oxidative stress);GO:0019904(protein domain specific binding);GO:0006366(transcription from RNA polymerase II promoter);GO:0043161(proteasome-mediated ubiquitin-dependent protein catabolic process);GO:0001102(RNA polymerase II activating transcription factor binding);GO:0010499(proteasomal ubiquitin-independent protein catabolic process);GO:1903206(negative regulation of hydrogen peroxide-induced cell death);GO:0071498(cellular response to fluid shear stress);GO:0071499(cellular response to laminar fluid shear stress);GO:1903071(positive regulation of ER-associated ubiquitin-dependent protein catabolic process);GO:0000976(transcription regulatory region sequence-specific DNA binding);GO:0010628(positive regulation of gene expression);GO:1902176(negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway);GO:0036499(PERK-mediated unfolded protein response);GO:0070301(cellular response to hydrogen peroxide);GO:0044212(transcription regulatory region DNA binding);GO:0036003(positive regulation of transcription from RNA polymerase II promoter in response to stress);GO:0032993(protein-DNA complex);GO:0030968(endoplasmic reticulum unfolded protein response);GO:0000980(RNA polymerase II distal enhancer sequence-specific DNA binding);GO:0001205(transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding);GO:0001221(transcription cofactor binding);GO:0008134(transcription factor binding);GO:0043565(sequence-specific DNA binding);GO:0006954(inflammatory response);GO:0007568(aging);GO:0010667(negative regulation of cardiac muscle cell apoptotic process);GO:0010976(positive regulation of neuron projection development);GO:0030194(positive regulation of blood coagulation);GO:0034976(response to endoplasmic reticulum stress);GO:0035690(cellular response to drug);GO:0036091(positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress);GO:0042149(cellular response to glucose starvation);GO:0043536(positive regulation of blood vessel endothelial cell migration);GO:0045454(cell redox homeostasis);GO:0045766(positive regulation of angiogenesis);GO:0045893(positive regulation of transcription, DNA-templated);GO:0045995(regulation of embryonic development);GO:0046223(aflatoxin catabolic process);GO:0046326(positive regulation of glucose import);GO:0060548(negative regulation of cell death);GO:0071456(cellular response to hypoxia);GO:1902037(negative regulation of hematopoietic stem cell differentiation);GO:1903788(positive regulation of glutathione biosynthetic process);GO:1904753(negative regulation of vascular associated smooth muscle cell migration);GO:2000121(regulation of removal of superoxide radicals);GO:2000379(positive regulation of reactive oxygen species metabolic process);GO:0000785(chromatin);GO:0000785(chromatin) | 04141(Protein processing in endoplasmic reticulum);05200(Pathways in cancer);05225(Hepatocellular carcinoma);05418(Fluid shear stress and atherosclerosis);05418(Fluid shear stress and atherosclerosis) | K05638 | NA | nuclear factor, erythroid 2 like 2 [Source:HGNC Symbol;Acc:HGNC:7782] | 12.4432937024586 | 0.58 | 21.3066164287635 | 4.41 | 4.05e-09 | 2.64e-06 | up | yes |
| ENSG00000258947 | TUBB3 | ENST00000554444;ENST00000553967;ENST00000554927;ENST00000555810;ENST00000315491;ENST00000555609;ENST00000556536;ENST00000554116 | GO:0005525(GTP binding);GO:0000166(nucleotide binding);GO:0003924(GTPase activity);GO:0015630(microtubule cytoskeleton);GO:0005200(structural constituent of cytoskeleton);GO:0007010(cytoskeleton organization);GO:0005874(microtubule);GO:0007017(microtubule-based process);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0070062(extracellular exosome);GO:0007411(axon guidance);GO:0030425(dendrite);GO:0030425(dendrite) | 04145(Phagosome);04540(Gap junction);05130(Pathogenic Escherichia coli infection);05130(Pathogenic Escherichia coli infection) | K07375 | NA | tubulin beta 3 class III [Source:HGNC Symbol;Acc:HGNC:20772] | 16.3056687151242 | 0.77 | 21.2331097349126 | 4.41 | 4.25e-09 | 2.65e-06 | up | yes |
| ENSG00000155008 | APOOL | ENST00000622540 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005739(mitochondrion);GO:0005743(mitochondrial inner membrane);GO:0005576(extracellular region);GO:0061617(MICOS complex);GO:0042407(cristae formation);GO:0005515(protein binding);GO:0002576(platelet degranulation);GO:0031093(platelet alpha granule lumen) | NA | NA | NA | apolipoprotein O like [Source:HGNC Symbol;Acc:HGNC:24009] | 0.09 | 2.15 | 0.04 | -4.559559736061 | 4.89e-09 | 2.93e-06 | down | yes |
| ENSG00000135482 | ZC3H10 | ENST00000257940;ENST00000551880 | GO:0003723(RNA binding);GO:0046872(metal ion binding);GO:0005515(protein binding);GO:0005515(protein binding) | NA | NA | NA | zinc finger CCCH-type containing 10 [Source:HGNC Symbol;Acc:HGNC:25893] | 1.96 | 0.09 | 21.200137153496 | 4.41 | 6.91e-09 | 3.97e-06 | up | yes |
| ENSG00000213644 | SAPCD2P1 | ENST00000395413 | NA | NA | NA | NA | suppressor APC domain containing 2 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:51277] | 0.48 | 10.831354 | 0.04 | -4.48601348915931 | 7.75e-09 | 4.28e-06 | down | yes |
| ENSG00000172340 | SUCLG2 | ENST00000492795;ENST00000493112;ENST00000307227 | GO:0046872(metal ion binding);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0006099(tricarboxylic acid cycle);GO:0005739(mitochondrion);GO:0003824(catalytic activity);GO:0005525(GTP binding);GO:0008152(metabolic process);GO:0005886(plasma membrane);GO:0016874(ligase activity);GO:0004776(succinate-CoA ligase (GDP-forming) activity);GO:0006104(succinyl-CoA metabolic process);GO:0005759(mitochondrial matrix);GO:0019003(GDP binding);GO:0046982(protein heterodimerization activity);GO:0006105(succinate metabolic process);GO:0043234(protein complex);GO:0045244(succinate-CoA ligase complex (GDP-forming));GO:0000287(magnesium ion binding);GO:0004775(succinate-CoA ligase (ADP-forming) activity);GO:0004775(succinate-CoA ligase (ADP-forming) activity) | 00020(Citrate cycle (TCA cycle));00640(Propanoate metabolism);00640(Propanoate metabolism) | K01900 | EC:6.2.1.4 6.2.1.5 | succinate-CoA ligase GDP-forming beta subunit [Source:HGNC Symbol;Acc:HGNC:11450] | 7.30 | 0.38 | 19.3258014102318 | 4.27 | 1.32e-08 | 7.04e-06 | up | yes |
| ENSG00000141985 | SH3GL1 | ENST00000269886;ENST00000598219;ENST00000593591 | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0042995(cell projection);GO:0005515(protein binding);GO:0045202(synapse);GO:0030054(cell junction);GO:0042802(identical protein binding);GO:0008289(lipid binding);GO:0005768(endosome);GO:0045296(cadherin binding);GO:0006897(endocytosis);GO:0002102(podosome);GO:0031901(early endosome membrane);GO:0007165(signal transduction);GO:0007417(central nervous system development);GO:0019902(phosphatase binding);GO:0051020(GTPase binding);GO:0016191(synaptic vesicle uncoating);GO:0016191(synaptic vesicle uncoating) | 04144(Endocytosis);04144(Endocytosis) | K11247 | NA | SH3 domain containing GRB2 like 1, endophilin A2 [Source:HGNC Symbol;Acc:HGNC:10830] | 26.0386755180461 | 1.43 | 18.2045394121924 | 4.19 | 1.57e-08 | 8.05e-06 | up | yes |
| ENSG00000253421 | ZNHIT1P1 | ENST00000523947 | NA | NA | NA | NA | zinc finger HIT-type containing 1 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:44474] | 51.756111 | 2.99 | 17.3250795353209 | 4.11 | 3.19e-08 | 1.58e-05 | up | yes |
| ENSG00000249489 | GAPDHP70 | ENST00000515281 | NA | NA | NA | NA | glyceraldehyde 3 phosphate dehydrogenase pseudogene 70 [Source:HGNC Symbol;Acc:HGNC:4148] | 29.962372 | 1.79 | 16.7592127820832 | 4.07 | 3.81e-08 | 1.77e-05 | up | yes |
| ENSG00000213742 | ZNF337-AS1 | ENST00000414393;ENST00000428254 | NA | NA | NA | NA | ZNF337 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:40759] | 0.14 | 2.50 | 0.06 | -4.18135211980167 | 3.83e-08 | 1.77e-05 | down | yes |
| ENSG00000175582 | RAB6A | ENST00000310653;ENST00000400470;ENST00000336083 | GO:0006810(transport);GO:0016020(membrane);GO:0015031(protein transport);GO:0005794(Golgi apparatus);GO:0005525(GTP binding);GO:0000166(nucleotide binding);GO:0003924(GTPase activity);GO:0005515(protein binding);GO:0000139(Golgi membrane);GO:0005886(plasma membrane);GO:0016192(vesicle-mediated transport);GO:0005789(endoplasmic reticulum membrane);GO:0005829(cytosol);GO:0070062(extracellular exosome);GO:0006890(retrograde vesicle-mediated transport, Golgi to ER);GO:0061024(membrane organization);GO:0019904(protein domain specific binding);GO:0016032(viral process);GO:0043312(neutrophil degranulation);GO:0032588(trans-Golgi network membrane);GO:0031489(myosin V binding);GO:0030667(secretory granule membrane);GO:0031410(cytoplasmic vesicle);GO:0019882(antigen processing and presentation);GO:0005802(trans-Golgi network);GO:0000042(protein targeting to Golgi);GO:0018125(peptidyl-cysteine methylation);GO:0034067(protein localization to Golgi apparatus);GO:0034498(early endosome to Golgi transport);GO:0072385(minus-end-directed organelle transport along microtubule);GO:0070381(endosome to plasma membrane transport vesicle);GO:0005622(intracellular);GO:0007264(small GTPase mediated signal transduction);GO:0007264(small GTPase mediated signal transduction) | NA | NA | NA | RAB6A, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:9786] | 14.0247224260621 | 0.85 | 16.5048354550396 | 4.04 | 3.94e-08 | 1.77e-05 | up | yes |
| ENSG00000213782 | DDX47 | ENST00000542123;ENST00000542832;ENST00000544497;ENST00000544032 | GO:0003676(nucleic acid binding);GO:0003723(RNA binding);GO:0006397(mRNA processing);GO:0008380(RNA splicing);GO:0005634(nucleus);GO:0016020(membrane);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0005730(nucleolus);GO:0016787(hydrolase activity);GO:0004386(helicase activity);GO:0005515(protein binding);GO:0006915(apoptotic process);GO:0006364(rRNA processing);GO:0005737(cytoplasm);GO:0005654(nucleoplasm);GO:0004004(ATP-dependent RNA helicase activity);GO:0010501(RNA secondary structure unwinding);GO:0008625(extrinsic apoptotic signaling pathway via death domain receptors);GO:0008625(extrinsic apoptotic signaling pathway via death domain receptors) | NA | NA | NA | DEAD-box helicase 47 [Source:HGNC Symbol;Acc:HGNC:18682] | 0.31 | 5.47 | 0.06 | -4.13352347120339 | 4.11e-08 | 1.78e-05 | down | yes |
| ENSG00000213585 | VDAC1 | ENST00000395044;ENST00000466080;ENST00000492324;ENST00000265333;ENST00000489906;ENST00000425992 | GO:0006810(transport);GO:0005634(nucleus);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0055085(transmembrane transport);GO:0008308(voltage-gated anion channel activity);GO:0098656(anion transmembrane transport);GO:1903959(regulation of anion transmembrane transport);GO:0005741(mitochondrial outer membrane);GO:0005739(mitochondrion);GO:0006811(ion transport);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0006915(apoptotic process);GO:0045121(membrane raft);GO:0006820(anion transport);GO:0070062(extracellular exosome);GO:0044325(ion channel binding);GO:0016032(viral process);GO:1903146(regulation of mitophagy);GO:0030855(epithelial cell differentiation);GO:0042645(mitochondrial nucleoid);GO:0015288(porin activity);GO:0016236(macroautophagy);GO:0019901(protein kinase binding);GO:0046930(pore complex);GO:0005253(anion channel activity);GO:0032403(protein complex binding);GO:0001662(behavioral fear response);GO:0006851(mitochondrial calcium ion transmembrane transport);GO:0007268(chemical synaptic transmission);GO:0007270(neuron-neuron synaptic transmission);GO:0007612(learning);GO:2000378(negative regulation of reactive oxygen species metabolic process);GO:0005743(mitochondrial inner membrane);GO:0008021(synaptic vesicle);GO:0043209(myelin sheath);GO:0043234(protein complex);GO:0043234(protein complex) | 04020(Calcium signaling pathway);04022(cGMP - PKG signaling pathway);04217(Necroptosis);04218(Cellular senescence);04621(NOD-like receptor signaling pathway);04979(Cholesterol metabolism);05012(Parkinson's disease);05016(Huntington's disease);05166(HTLV-I infection);05164(Influenza A);05164(Influenza A) | K05862 | NA | voltage dependent anion channel 1 [Source:HGNC Symbol;Acc:HGNC:12669] | 3.07 | 50.5145428510907 | 0.06 | -4.03987368910958 | 5.68e-08 | 2.40e-05 | down | yes |
| ENSG00000083312 | TNPO1 | ENST00000337273;ENST00000605210;ENST00000511754;ENST00000513944 | GO:0003723(RNA binding);GO:0006810(transport);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0006886(intracellular protein transport);GO:0015031(protein transport);GO:0005515(protein binding);GO:0008536(Ran GTPase binding);GO:0035735(intraciliary transport involved in cilium assembly);GO:0005929(cilium);GO:0005829(cytosol);GO:0070062(extracellular exosome);GO:0043488(regulation of mRNA stability);GO:0016032(viral process);GO:0008565(protein transporter activity);GO:0006607(NLS-bearing protein import into nucleus);GO:0031965(nuclear membrane);GO:0034399(nuclear periphery);GO:0008139(nuclear localization sequence binding);GO:0006610(ribosomal protein import into nucleus);GO:0000060(protein import into nucleus, translocation);GO:0000060(protein import into nucleus, translocation) | NA | NA | NA | transportin 1 [Source:HGNC Symbol;Acc:HGNC:6401] | 0.28 | 4.42 | 0.06 | -3.99427017188277 | 8.41e-08 | 3.45e-05 | down | yes |
| ENSG00000099860 | GADD45B | ENST00000592937;ENST00000586759 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0005515(protein binding);GO:0006915(apoptotic process);GO:0007275(multicellular organism development);GO:0030154(cell differentiation);GO:0051726(regulation of cell cycle);GO:0006950(response to stress);GO:0043065(positive regulation of apoptotic process);GO:0046330(positive regulation of JNK cascade);GO:0000185(activation of MAPKKK activity);GO:0000186(activation of MAPKK activity);GO:0006469(negative regulation of protein kinase activity);GO:1900745(positive regulation of p38MAPK cascade);GO:1900745(positive regulation of p38MAPK cascade) | 04010(MAPK signaling pathway);04064(NF-kappa B signaling pathway);04068(FoxO signaling pathway);04110(Cell cycle);04210(Apoptosis);04115(p53 signaling pathway);04218(Cellular senescence);05200(Pathways in cancer);05202(Transcriptional misregulation in cancers);05210(Colorectal cancer);05212(Pancreatic cancer);05225(Hepatocellular carcinoma);05226(Gastric cancer);05214(Glioma);05216(Thyroid cancer);05220(Chronic myeloid leukemia);05217(Basal cell carcinoma);05218(Melanoma);05213(Endometrial cancer);05224(Breast cancer);05222(Small cell lung cancer);05223(Non-small cell lung cancer);05223(Non-small cell lung cancer) | NA | NA | growth arrest and DNA damage inducible beta [Source:HGNC Symbol;Acc:HGNC:4096] | 0.56 | 8.51 | 0.07 | -3.93866268688729 | 1.45e-07 | 5.81e-05 | down | yes |
| ENSG00000163866 | SMIM12 | ENST00000446026;ENST00000426886;ENST00000417239;ENST00000521580 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0016021(integral component of membrane) | NA | NA | NA | small integral membrane protein 12 [Source:HGNC Symbol;Acc:HGNC:25154] | 0.46 | 6.70 | 0.07 | -3.8623420945615 | 1.84e-07 | 7.17e-05 | down | yes |
| ENSG00000226789 | AC110926.1 | ENST00000412499 | NA | NA | NA | NA | 3.35 | 0.23 | 14.6066617875129 | 3.87 | 2.00e-07 | 7.39e-05 | up | yes | |
| ENSG00000197119 | SLC25A29 | ENST00000553574;ENST00000556844;ENST00000554912;ENST00000359232;ENST00000557438 | GO:0006810(transport);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0055085(transmembrane transport);GO:0005743(mitochondrial inner membrane);GO:0005739(mitochondrion);GO:1990822(basic amino acid transmembrane transport);GO:1903826(arginine transmembrane transport);GO:0015171(amino acid transmembrane transporter activity);GO:0006839(mitochondrial transport);GO:1902616(acyl carnitine transmembrane transport);GO:0006865(amino acid transport);GO:0015174(basic amino acid transmembrane transporter activity);GO:0005289(high-affinity arginine transmembrane transporter activity);GO:0005292(high-affinity lysine transmembrane transporter activity);GO:0015227(acyl carnitine transmembrane transporter activity);GO:0006844(acyl carnitine transport);GO:0015822(ornithine transport);GO:0089709(L-histidine transmembrane transport);GO:1903400(L-arginine transmembrane transport);GO:1903401(L-lysine transmembrane transport);GO:1990575(mitochondrial L-ornithine transmembrane transport);GO:1990575(mitochondrial L-ornithine transmembrane transport) | NA | NA | NA | solute carrier family 25 member 29 [Source:HGNC Symbol;Acc:HGNC:20116] | 0.57 | 8.22 | 0.07 | -3.84457565324725 | 2.00e-07 | 7.39e-05 | down | yes |
| ENSG00000102401 | ARMCX3 | ENST00000477980;ENST00000341189 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0034613(cellular protein localization);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0031307(integral component of mitochondrial outer membrane);GO:0031307(integral component of mitochondrial outer membrane) | NA | NA | NA | armadillo repeat containing, X-linked 3 [Source:HGNC Symbol;Acc:HGNC:24065] | 7.26 | 0.53 | 13.6688369498994 | 3.77 | 2.28e-07 | 8.19e-05 | up | yes |
| ENSG00000128609 | NDUFA5 | ENST00000471770;ENST00000466896;ENST00000467117;ENST00000490618 | GO:0016020(membrane);GO:0055114(oxidation-reduction process);GO:0005743(mitochondrial inner membrane);GO:0005739(mitochondrion);GO:0005515(protein binding);GO:0070469(respiratory chain);GO:0016651(oxidoreductase activity, acting on NAD(P)H);GO:0022904(respiratory electron transport chain);GO:0032981(mitochondrial respiratory chain complex I assembly);GO:0008137(NADH dehydrogenase (ubiquinone) activity);GO:0006120(mitochondrial electron transport, NADH to ubiquinone);GO:0005747(mitochondrial respiratory chain complex I);GO:0045271(respiratory chain complex I);GO:0045271(respiratory chain complex I) | 00190(Oxidative phosphorylation);04723(Retrograde endocannabinoid signaling);05010(Alzheimer's disease);05012(Parkinson's disease);05016(Huntington's disease);04932(Non-alcoholic fatty liver disease (NAFLD));04932(Non-alcoholic fatty liver disease (NAFLD)) | K03949 | NA | NADH:ubiquinone oxidoreductase subunit A5 [Source:HGNC Symbol;Acc:HGNC:7688] | 1.03 | 0.07 | 14.5403950564581 | 3.86 | 2.43e-07 | 8.53e-05 | up | yes |
| ENSG00000160131 | VMA21 | ENST00000330374;ENST00000370361;ENST00000477649 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0031410(cytoplasmic vesicle);GO:0070072(vacuolar proton-transporting V-type ATPase complex assembly);GO:0033116(endoplasmic reticulum-Golgi intermediate compartment membrane);GO:0012507(ER to Golgi transport vesicle membrane);GO:0043462(regulation of ATPase activity);GO:0005764(lysosome);GO:0005764(lysosome) | NA | NA | NA | VMA21, vacuolar ATPase assembly factor [Source:HGNC Symbol;Acc:HGNC:22082] | 0.50 | 6.96 | 0.07 | -3.79579686141496 | 2.88e-07 | 9.86e-05 | down | yes |
| ENSG00000254413 | CHKB-CPT1B | ENST00000492556;ENST00000453634 | GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0046474(glycerophospholipid biosynthetic process);GO:0046474(glycerophospholipid biosynthetic process) | NA | NA | NA | CHKB-CPT1B readthrough (NMD candidate) [Source:HGNC Symbol;Acc:HGNC:41998] | 2.34 | 0.18 | 13.3014946372298 | 3.73 | 3.87e-07 | 0.00 | up | yes |
| ENSG00000152944 | MED21 | ENST00000282892;ENST00000544998;ENST00000536711 | GO:0005634(nucleus);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0005515(protein binding);GO:0061630(ubiquitin protein ligase activity);GO:0001824(blastocyst development);GO:0016567(protein ubiquitination);GO:0019827(stem cell population maintenance);GO:0000151(ubiquitin ligase complex);GO:0016592(mediator complex);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0006357(regulation of transcription from RNA polymerase II promoter);GO:0003899(DNA-directed 5'-3' RNA polymerase activity);GO:0003713(transcription coactivator activity);GO:0001104(RNA polymerase II transcription cofactor activity);GO:0001104(RNA polymerase II transcription cofactor activity) | NA | NA | NA | mediator complex subunit 21 [Source:HGNC Symbol;Acc:HGNC:11473] | 6.60 | 0.52 | 12.5886470377455 | 3.65 | 5.04e-07 | 0.00 | up | yes |
| ENSG00000185515 | BRCC3 | ENST00000340647;ENST00000369462 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0005654(nucleoplasm);GO:0004843(thiol-dependent ubiquitin-specific protease activity);GO:0016787(hydrolase activity);GO:0016569(covalent chromatin modification);GO:0051301(cell division);GO:0050790(regulation of catalytic activity);GO:0006281(DNA repair);GO:0005856(cytoskeleton);GO:0008233(peptidase activity);GO:0006508(proteolysis);GO:0006974(cellular response to DNA damage stimulus);GO:0007049(cell cycle);GO:0036459(thiol-dependent ubiquitinyl hydrolase activity);GO:0008237(metallopeptidase activity);GO:0070531(BRCA1-A complex);GO:0070552(BRISC complex);GO:0000922(spindle pole);GO:0070536(protein K63-linked deubiquitination);GO:0005515(protein binding);GO:0005829(cytosol);GO:0016579(protein deubiquitination);GO:0061578(Lys63-specific deubiquitinase activity);GO:0031572(G2 DNA damage checkpoint);GO:0045739(positive regulation of DNA repair);GO:0006302(double-strand break repair);GO:0006303(double-strand break repair via nonhomologous end joining);GO:0010212(response to ionizing radiation);GO:0031593(polyubiquitin binding);GO:0030234(enzyme regulator activity);GO:0010165(response to X-ray);GO:0070537(histone H2A K63-linked deubiquitination);GO:0000151(ubiquitin ligase complex);GO:0000152(nuclear ubiquitin ligase complex);GO:0000152(nuclear ubiquitin ligase complex) | 03440(Homologous recombination);04621(NOD-like receptor signaling pathway);04621(NOD-like receptor signaling pathway) | K11864 | EC:3.4.19.- | BRCA1/BRCA2-containing complex subunit 3 [Source:HGNC Symbol;Acc:HGNC:24185] | 0.42 | 5.37 | 0.08 | -3.68772460241875 | 6.32e-07 | 0.00 | down | yes |
| ENSG00000183955 | KMT5A | ENST00000537270;ENST00000461103;ENST00000478781;ENST00000437502;ENST00000402868 | GO:0005634(nucleus);GO:0005829(cytosol);GO:0008168(methyltransferase activity);GO:0016740(transferase activity);GO:0032259(methylation);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0016569(covalent chromatin modification);GO:0051301(cell division);GO:0005694(chromosome);GO:0005515(protein binding);GO:0007049(cell cycle);GO:0018024(histone-lysine N-methyltransferase activity);GO:0018026(peptidyl-lysine monomethylation);GO:0016279(protein-lysine N-methyltransferase activity);GO:0034968(histone lysine methylation);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0003714(transcription corepressor activity);GO:0005654(nucleoplasm);GO:0045892(negative regulation of transcription, DNA-templated);GO:1901796(regulation of signal transduction by p53 class mediator);GO:0002039(p53 binding);GO:0042799(histone methyltransferase activity (H4-K20 specific));GO:0016278(lysine N-methyltransferase activity);GO:0034770(histone H4-K20 methylation);GO:0043516(regulation of DNA damage response, signal transduction by p53 class mediator);GO:0043516(regulation of DNA damage response, signal transduction by p53 class mediator) | 00310(Lysine degradation);00310(Lysine degradation) | K11428 | EC:2.1.1.43 | lysine methyltransferase 5A [Source:HGNC Symbol;Acc:HGNC:29489] | 6.03 | 0.52 | 11.7049049497019 | 3.55 | 8.48e-07 | 0.00 | up | yes |
| ENSG00000198633 | ZNF534 | ENST00000301085 | GO:0003676(nucleic acid binding);GO:0006355(regulation of transcription, DNA-templated);GO:0005622(intracellular);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0006351(transcription, DNA-templated);GO:0003677(DNA binding);GO:0003700(transcription factor activity, sequence-specific DNA binding) | NA | NA | NA | zinc finger protein 534 [Source:HGNC Symbol;Acc:HGNC:26337] | 0.34 | 4.20 | 0.08 | -3.62638198412686 | 9.65e-07 | 0.00 | down | yes |
| ENSG00000170545 | SMAGP | ENST00000603798;ENST00000380103;ENST00000603838 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005654(nucleoplasm);GO:0005886(plasma membrane);GO:0030054(cell junction);GO:0005515(protein binding);GO:0030659(cytoplasmic vesicle membrane);GO:0031410(cytoplasmic vesicle);GO:0005887(integral component of plasma membrane);GO:0004872(receptor activity);GO:0005102(receptor binding);GO:0042803(protein homodimerization activity);GO:0050839(cell adhesion molecule binding);GO:0007156(homophilic cell adhesion via plasma membrane adhesion molecules);GO:0007157(heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules);GO:0005913(cell-cell adherens junction);GO:0008037(cell recognition);GO:0008037(cell recognition) | NA | NA | NA | small cell adhesion glycoprotein [Source:HGNC Symbol;Acc:HGNC:26918] | 5.76 | 0.51 | 11.2304263337855 | 3.49 | 1.30e-06 | 0.00 | up | yes |
| ENSG00000138107 | ACTR1A | ENST00000369905;ENST00000636707;ENST00000480947 | GO:0005869(dynactin complex);GO:0005737(cytoplasm);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0015630(microtubule cytoskeleton);GO:0005856(cytoskeleton);GO:0005813(centrosome);GO:0005815(microtubule organizing center);GO:0005515(protein binding);GO:0005829(cytosol);GO:0070062(extracellular exosome);GO:0005938(cell cortex);GO:0000086(G2/M transition of mitotic cell cycle);GO:0010389(regulation of G2/M transition of mitotic cell cycle);GO:0097711(ciliary basal body docking);GO:0019886(antigen processing and presentation of exogenous peptide antigen via MHC class II);GO:0016192(vesicle-mediated transport);GO:0006888(ER to Golgi vesicle-mediated transport);GO:0005875(microtubule associated complex);GO:0099738(cell cortex region);GO:0007283(spermatogenesis);GO:0002177(manchette);GO:0005814(centriole);GO:0030137(COPI-coated vesicle);GO:0043209(myelin sheath);GO:0043209(myelin sheath) | NA | NA | NA | ARP1 actin related protein 1 homolog A [Source:HGNC Symbol;Acc:HGNC:167] | 49.5591658816072 | 4.62 | 10.7167659123106 | 3.42 | 1.52e-06 | 0.00 | up | yes |
| ENSG00000179085 | DPM3 | ENST00000368399 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0006486(protein glycosylation);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0005515(protein binding);GO:0006506(GPI anchor biosynthetic process);GO:0031647(regulation of protein stability);GO:0005975(carbohydrate metabolic process);GO:0004582(dolichyl-phosphate beta-D-mannosyltransferase activity);GO:0018279(protein N-linked glycosylation via asparagine);GO:0035268(protein mannosylation);GO:0035269(protein O-linked mannosylation);GO:0033185(dolichol-phosphate-mannose synthase complex);GO:0030176(integral component of endoplasmic reticulum membrane);GO:0018406(protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan);GO:0031501(mannosyltransferase complex) | 00510(N-Glycan biosynthesis) | K09659 | NA | dolichyl-phosphate mannosyltransferase subunit 3 [Source:HGNC Symbol;Acc:HGNC:3007] | 0.95 | 11.6041780418006 | 0.08 | -3.61116013453559 | 1.57e-06 | 0.00 | down | yes |
| ENSG00000178458 | H3F3AP6 | ENST00000230495 | NA | NA | NA | NA | H3 histone, family 3A, pseudogene 6 [Source:HGNC Symbol;Acc:HGNC:42982] | 4.32 | 49.419357 | 0.09 | -3.51466984214763 | 1.65e-06 | 0.00 | down | yes |
| ENSG00000167625 | ZNF526 | ENST00000597945;ENST00000301215 | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0003677(DNA binding) | NA | NA | NA | zinc finger protein 526 [Source:HGNC Symbol;Acc:HGNC:29415] | 4.85 | 0.45 | 10.8806102588578 | 3.44 | 1.66e-06 | 0.00 | up | yes |
| ENSG00000179195 | ZNF664 | ENST00000539644;ENST00000542820;ENST00000539501;ENST00000540498;ENST00000537532;ENST00000538932;ENST00000542493 | GO:0005634(nucleus);GO:0003676(nucleic acid binding);GO:0046872(metal ion binding);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0003674(molecular_function);GO:0008150(biological_process);GO:0008150(biological_process) | NA | NA | NA | zinc finger protein 664 [Source:HGNC Symbol;Acc:HGNC:25406] | 2.53 | 27.6254241755521 | 0.09 | -3.44822950028758 | 1.97e-06 | 0.00 | down | yes |
| ENSG00000050344 | NFE2L3 | ENST00000056233 | GO:0005634(nucleus);GO:0006355(regulation of transcription, DNA-templated);GO:0006351(transcription, DNA-templated);GO:0003677(DNA binding);GO:0006357(regulation of transcription from RNA polymerase II promoter);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0005515(protein binding);GO:0005737(cytoplasm);GO:0003713(transcription coactivator activity);GO:0006366(transcription from RNA polymerase II promoter);GO:0045893(positive regulation of transcription, DNA-templated);GO:0000976(transcription regulatory region sequence-specific DNA binding);GO:0000978(RNA polymerase II core promoter proximal region sequence-specific DNA binding);GO:0001078(transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding);GO:0000122(negative regulation of transcription from RNA polymerase II promoter) | NA | NA | NA | nuclear factor, erythroid 2 like 3 [Source:HGNC Symbol;Acc:HGNC:7783] | 1.54 | 16.7828741614498 | 0.09 | -3.44649211349937 | 2.07e-06 | 0.00 | down | yes |
| ENSG00000284057 | AP001273.2 | ENST00000638767 | GO:0005634(nucleus);GO:0001104(RNA polymerase II transcription cofactor activity);GO:0006357(regulation of transcription from RNA polymerase II promoter);GO:0016592(mediator complex) | NA | NA | NA | 2.70 | 0.25 | 11.0046101169853 | 3.46 | 2.31e-06 | 0.00 | up | yes | |
| ENSG00000125814 | NAPB | ENST00000617876;ENST00000487502;ENST00000398425 | GO:0006810(transport);GO:0016020(membrane);GO:0006886(intracellular protein transport);GO:0015031(protein transport);GO:0005515(protein binding);GO:0016192(vesicle-mediated transport);GO:0019905(syntaxin binding);GO:0070062(extracellular exosome);GO:0031201(SNARE complex);GO:0061025(membrane fusion);GO:0005483(soluble NSF attachment protein activity);GO:0035494(SNARE complex disassembly);GO:0005774(vacuolar membrane);GO:0010807(regulation of synaptic vesicle priming);GO:0035249(synaptic transmission, glutamatergic);GO:0043209(myelin sheath);GO:0070044(synaptobrevin 2-SNAP-25-syntaxin-1a complex);GO:0070044(synaptobrevin 2-SNAP-25-syntaxin-1a complex) | NA | NA | NA | NSF attachment protein beta [Source:HGNC Symbol;Acc:HGNC:15751] | 1.87 | 0.17 | 10.725466604895 | 3.42 | 2.38e-06 | 0.00 | up | yes |
| ENSG00000176014 | TUBB6 | ENST00000586810;ENST00000591909;ENST00000590693;ENST00000417736;ENST00000590967;ENST00000317702;ENST00000587204 | GO:0005525(GTP binding);GO:0003924(GTPase activity);GO:0005200(structural constituent of cytoskeleton);GO:0007010(cytoskeleton organization);GO:0005874(microtubule);GO:0007017(microtubule-based process);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0000166(nucleotide binding);GO:0015630(microtubule cytoskeleton);GO:0005856(cytoskeleton);GO:0003674(molecular_function);GO:0070062(extracellular exosome);GO:0070062(extracellular exosome) | 04145(Phagosome);04540(Gap junction);05130(Pathogenic Escherichia coli infection);05130(Pathogenic Escherichia coli infection) | K07375 | NA | tubulin beta 6 class V [Source:HGNC Symbol;Acc:HGNC:20776] | 56.8556907393733 | 5.71 | 9.96 | 3.32 | 2.83e-06 | 0.00 | up | yes |
| ENSG00000154102 | C16orf74 | ENST00000602719;ENST00000602583;ENST00000602758 | GO:0005515(protein binding);GO:0005515(protein binding) | NA | NA | NA | chromosome 16 open reading frame 74 [Source:HGNC Symbol;Acc:HGNC:23362] | 7.88 | 0.76 | 10.3378931526309 | 3.37 | 2.84e-06 | 0.00 | up | yes |
| ENSG00000235173 | HGH1 | ENST00000525101;ENST00000530409;ENST00000347708 | GO:0003674(molecular_function);GO:0008150(biological_process);GO:0005575(cellular_component);GO:0005575(cellular_component) | NA | NA | NA | HGH1 homolog [Source:HGNC Symbol;Acc:HGNC:24161] | 23.586992934968 | 2.37 | 9.96 | 3.32 | 2.98e-06 | 0.00 | up | yes |
| ENSG00000062485 | CS | ENST00000549143;ENST00000546585;ENST00000542324;ENST00000551473;ENST00000546621;ENST00000551137 | GO:0016740(transferase activity);GO:0006099(tricarboxylic acid cycle);GO:0005739(mitochondrion);GO:0046912(transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer);GO:0006101(citrate metabolic process);GO:0004108(citrate (Si)-synthase activity);GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005759(mitochondrial matrix);GO:0005975(carbohydrate metabolic process);GO:0070062(extracellular exosome);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0016021(integral component of membrane) | 00020(Citrate cycle (TCA cycle));00630(Glyoxylate and dicarboxylate metabolism);00630(Glyoxylate and dicarboxylate metabolism) | NA | NA | citrate synthase [Source:HGNC Symbol;Acc:HGNC:2422] | 19.7775783570759 | 2.00 | 9.91 | 3.31 | 3.02e-06 | 0.00 | up | yes |
| ENSG00000204628 | RACK1 | ENST00000514455;ENST00000514183;ENST00000508044;ENST00000507261;ENST00000510199;ENST00000507000;ENST00000512805;ENST00000511473 | GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0016567(protein ubiquitination);GO:0030529(intracellular ribonucleoprotein complex);GO:0005840(ribosome);GO:0005739(mitochondrion);GO:0005886(plasma membrane);GO:0042995(cell projection);GO:0005515(protein binding);GO:0007049(cell cycle);GO:0048511(rhythmic process);GO:0006915(apoptotic process);GO:0007275(multicellular organism development);GO:0048471(perinuclear region of cytoplasm);GO:0065009(regulation of molecular function);GO:0043022(ribosome binding);GO:0030425(dendrite);GO:0030496(midbody);GO:0045296(cadherin binding);GO:2000114(regulation of establishment of cell polarity);GO:0043204(perikaryon);GO:0070062(extracellular exosome);GO:0001934(positive regulation of protein phosphorylation);GO:0001891(phagocytic cup);GO:0043547(positive regulation of GTPase activity);GO:0042169(SH2 domain binding);GO:0040008(regulation of growth);GO:0032947(protein complex scaffold);GO:0006417(regulation of translation);GO:0030178(negative regulation of Wnt signaling pathway);GO:0030335(positive regulation of cell migration);GO:0043065(positive regulation of apoptotic process);GO:0016032(viral process);GO:0005102(receptor binding);GO:0017148(negative regulation of translation);GO:0008200(ion channel inhibitor activity);GO:0006412(translation);GO:0043473(pigmentation);GO:1903208(negative regulation of hydrogen peroxide-induced neuron death);GO:0043005(neuron projection);GO:0043025(neuronal cell body);GO:0044297(cell body);GO:0010629(negative regulation of gene expression);GO:0019903(protein phosphatase binding);GO:0030308(negative regulation of cell growth);GO:0042803(protein homodimerization activity);GO:0051726(regulation of cell cycle);GO:2001244(positive regulation of intrinsic apoptotic signaling pathway);GO:0030971(receptor tyrosine kinase binding);GO:0051302(regulation of cell division);GO:0019899(enzyme binding);GO:0008656(cysteine-type endopeptidase activator activity involved in apoptotic process);GO:0006919(activation of cysteine-type endopeptidase activity involved in apoptotic process);GO:0051898(negative regulation of protein kinase B signaling);GO:0071333(cellular response to glucose stimulus);GO:0010803(regulation of tumor necrosis factor-mediated signaling pathway);GO:0071363(cellular response to growth factor stimulus);GO:0032880(regulation of protein localization);GO:0033137(negative regulation of peptidyl-serine phosphorylation);GO:0050765(negative regulation of phagocytosis);GO:2000304(positive regulation of ceramide biosynthetic process);GO:0032436(positive regulation of proteasomal ubiquitin-dependent protein catabolic process);GO:1990630(IRE1-RACK1-PP2A complex);GO:0030822(positive regulation of cAMP catabolic process);GO:0042998(positive regulation of Golgi to plasma membrane protein transport);GO:0030292(protein tyrosine kinase inhibitor activity);GO:0061099(negative regulation of protein tyrosine kinase activity);GO:0007369(gastrulation);GO:0005080(protein kinase C binding);GO:0032464(positive regulation of protein homooligomerization);GO:2000543(positive regulation of gastrulation);GO:0015935(small ribosomal subunit);GO:0035591(signaling adaptor activity);GO:0051343(positive regulation of cyclic-nucleotide phosphodiesterase activity);GO:0051901(positive regulation of mitochondrial depolarization);GO:0072344(rescue of stalled ribosome);GO:1900102(negative regulation of endoplasmic reticulum unfolded protein response);GO:1900102(negative regulation of endoplasmic reticulum unfolded protein response) | 05162(Measles);05162(Measles) | K14753 | NA | receptor for activated C kinase 1 [Source:HGNC Symbol;Acc:HGNC:4399] | 4.36 | 45.0407227111853 | 0.10 | -3.36916115631552 | 3.11e-06 | 0.00 | down | yes |
| ENSG00000105193 | RPS16 | ENST00000602153;ENST00000339471;ENST00000601390;ENST00000595386 | GO:0030529(intracellular ribonucleoprotein complex);GO:0003735(structural constituent of ribosome);GO:0006412(translation);GO:0005840(ribosome);GO:0003723(RNA binding);GO:0016020(membrane);GO:0005515(protein binding);GO:0022627(cytosolic small ribosomal subunit);GO:0005925(focal adhesion);GO:0006364(rRNA processing);GO:0042274(ribosomal small subunit biogenesis);GO:0000462(maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA));GO:0005654(nucleoplasm);GO:0005829(cytosol);GO:0070062(extracellular exosome);GO:0031012(extracellular matrix);GO:0006413(translational initiation);GO:0000184(nuclear-transcribed mRNA catabolic process, nonsense-mediated decay);GO:0019083(viral transcription);GO:0006614(SRP-dependent cotranslational protein targeting to membrane);GO:0097421(liver regeneration);GO:1990830(cellular response to leukemia inhibitory factor);GO:0015935(small ribosomal subunit);GO:0015935(small ribosomal subunit) | 03010(Ribosome);03010(Ribosome) | K02960 | NA | ribosomal protein S16 [Source:HGNC Symbol;Acc:HGNC:10396] | 7.05 | 0.70 | 10.1036348330618 | 3.34 | 3.30e-06 | 0.00 | up | yes |
| ENSG00000129128 | SPCS3 | ENST00000507001;ENST00000507678;ENST00000503362 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0016787(hydrolase activity);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0008233(peptidase activity);GO:0006508(proteolysis);GO:0006465(signal peptide processing);GO:0005787(signal peptidase complex);GO:0043231(intracellular membrane-bounded organelle);GO:0031090(organelle membrane);GO:0045047(protein targeting to ER);GO:0045047(protein targeting to ER) | 03060(Protein export);03060(Protein export) | K12948 | EC:3.4.-.- | signal peptidase complex subunit 3 [Source:HGNC Symbol;Acc:HGNC:26212] | 0.41 | 4.29 | 0.10 | -3.38109458959055 | 3.36e-06 | 0.00 | down | yes |
| ENSG00000142686 | C1orf216 | ENST00000503824;ENST00000270815 | GO:0005515(protein binding);GO:0005515(protein binding) | NA | NA | NA | chromosome 1 open reading frame 216 [Source:HGNC Symbol;Acc:HGNC:26800] | 0.48 | 5.03 | 0.10 | -3.37876300528418 | 3.70e-06 | 0.00 | down | yes |
| ENSG00000166548 | TK2 | ENST00000299697;ENST00000562552;ENST00000563478;ENST00000562484 | GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0016740(transferase activity);GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0006139(nucleobase-containing compound metabolic process);GO:0005739(mitochondrion);GO:0019206(nucleoside kinase activity);GO:0009165(nucleotide biosynthetic process);GO:0005759(mitochondrial matrix);GO:0009157(deoxyribonucleoside monophosphate biosynthetic process);GO:0004797(thymidine kinase activity);GO:0071897(DNA biosynthetic process);GO:0043097(pyrimidine nucleoside salvage);GO:0004137(deoxycytidine kinase activity);GO:0046092(deoxycytidine metabolic process);GO:0046104(thymidine metabolic process);GO:0046104(thymidine metabolic process) | 00240(Pyrimidine metabolism);00983(Drug metabolism - other enzymes);00983(Drug metabolism - other enzymes) | K00857 | EC:2.7.1.21 | thymidine kinase 2, mitochondrial [Source:HGNC Symbol;Acc:HGNC:11831] | 4.40 | 0.45 | 9.76 | 3.29 | 3.88e-06 | 0.00 | up | yes |
| ENSG00000168000 | BSCL2 | ENST00000463679;ENST00000403550;ENST00000449636;ENST00000531524;ENST00000413908;ENST00000530009;ENST00000405837 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0019915(lipid storage);GO:0006629(lipid metabolic process);GO:0016042(lipid catabolic process);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0005515(protein binding);GO:0003674(molecular_function);GO:0045444(fat cell differentiation);GO:0030176(integral component of endoplasmic reticulum membrane);GO:0034389(lipid particle organization);GO:0050995(negative regulation of lipid catabolic process);GO:0050995(negative regulation of lipid catabolic process) | NA | NA | NA | BSCL2, seipin lipid droplet biogenesis associated [Source:HGNC Symbol;Acc:HGNC:15832] | 0.87 | 8.77 | 0.10 | -3.33666917835324 | 4.24e-06 | 0.00 | down | yes |
| ENSG00000188242 | PP7080 | ENST00000342584;ENST00000510714 | NA | NA | NA | NA | uncharacterized LOC25845 [Source:NCBI gene;Acc:25845] | 5.98 | 0.61 | 9.78 | 3.29 | 4.24e-06 | 0.00 | up | yes |
| ENSG00000169567 | HINT1 | ENST00000511475;ENST00000304043;ENST00000513345;ENST00000506207 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0000166(nucleotide binding);GO:0005829(cytosol);GO:0016787(hydrolase activity);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003824(catalytic activity);GO:0005886(plasma membrane);GO:0006915(apoptotic process);GO:0070062(extracellular exosome);GO:0000118(histone deacetylase complex);GO:0007165(signal transduction);GO:0005856(cytoskeleton);GO:0072332(intrinsic apoptotic signaling pathway by p53 class mediator);GO:0005080(protein kinase C binding);GO:0009154(purine ribonucleotide catabolic process);GO:0050850(positive regulation of calcium-mediated signaling);GO:0050850(positive regulation of calcium-mediated signaling) | NA | NA | NA | histidine triad nucleotide binding protein 1 [Source:HGNC Symbol;Acc:HGNC:4912] | 1.49 | 15.0924170578512 | 0.10 | -3.3389235089633 | 4.36e-06 | 0.00 | down | yes |
| ENSG00000113558 | SKP1 | ENST00000522552;ENST00000520417;ENST00000523966;ENST00000353411 | GO:0016567(protein ubiquitination);GO:0005515(protein binding);GO:0006511(ubiquitin-dependent protein catabolic process);GO:0038095(Fc-epsilon receptor signaling pathway);GO:0005654(nucleoplasm);GO:0005829(cytosol);GO:0070062(extracellular exosome);GO:0000209(protein polyubiquitination);GO:0002223(stimulatory C-type lectin receptor signaling pathway);GO:0010972(negative regulation of G2/M transition of mitotic cell cycle);GO:0031146(SCF-dependent proteasomal ubiquitin-dependent protein catabolic process);GO:0038061(NIK/NF-kappaB signaling);GO:0043161(proteasome-mediated ubiquitin-dependent protein catabolic process);GO:0043687(post-translational protein modification);GO:0050852(T cell receptor signaling pathway);GO:0051437(positive regulation of ubiquitin-protein ligase activity involved in regulation of mitotic cell cycle transition);GO:0000086(G2/M transition of mitotic cell cycle);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0004842(ubiquitin-protein transferase activity);GO:0031519(PcG protein complex);GO:0019904(protein domain specific binding);GO:0016055(Wnt signaling pathway);GO:0051403(stress-activated MAPK cascade);GO:0008013(beta-catenin binding);GO:0061630(ubiquitin protein ligase activity);GO:0097602(cullin family protein binding);GO:0035518(histone H2A monoubiquitination);GO:0006879(cellular iron ion homeostasis);GO:0019005(SCF ubiquitin ligase complex);GO:0010265(SCF complex assembly);GO:0031467(Cul7-RING ubiquitin ligase complex);GO:0051457(maintenance of protein location in nucleus);GO:0051457(maintenance of protein location in nucleus) | 04141(Protein processing in endoplasmic reticulum);04120(Ubiquitin mediated proteolysis);04310(Wnt signaling pathway);04350(TGF-beta signaling pathway);04110(Cell cycle);04114(Oocyte meiosis);04710(Circadian rhythm);05200(Pathways in cancer);05168(Herpes simplex infection);05168(Herpes simplex infection) | K03094 | NA | S-phase kinase associated protein 1 [Source:HGNC Symbol;Acc:HGNC:10899] | 0.20 | 2.04 | 0.10 | -3.32354663165017 | 4.64e-06 | 0.00 | down | yes |
| ENSG00000240303 | ACAD11 | ENST00000496418;ENST00000487024 | GO:0005634(nucleus);GO:0016491(oxidoreductase activity);GO:0050660(flavin adenine dinucleotide binding);GO:0055114(oxidation-reduction process);GO:0016627(oxidoreductase activity, acting on the CH-CH group of donors);GO:0008152(metabolic process);GO:0005739(mitochondrion);GO:0005777(peroxisome);GO:0031966(mitochondrial membrane);GO:0005743(mitochondrial inner membrane);GO:0006635(fatty acid beta-oxidation);GO:0003995(acyl-CoA dehydrogenase activity);GO:0033539(fatty acid beta-oxidation using acyl-CoA dehydrogenase);GO:0004466(long-chain-acyl-CoA dehydrogenase activity);GO:0017099(very-long-chain-acyl-CoA dehydrogenase activity);GO:0070991(medium-chain-acyl-CoA dehydrogenase activity);GO:0070991(medium-chain-acyl-CoA dehydrogenase activity) | NA | NA | NA | acyl-CoA dehydrogenase family member 11 [Source:HGNC Symbol;Acc:HGNC:30211] | 0.13 | 1.32 | 0.09 | -3.39680476616964 | 4.83e-06 | 0.00 | down | yes |
| ENSG00000126005 | MMP24-AS1 | ENST00000435366;ENST00000566203;ENST00000438751;ENST00000594413;ENST00000456350 | NA | NA | NA | NA | MMP24 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:44421] | 0.41 | 4.14 | 0.10 | -3.32526719332005 | 4.87e-06 | 0.00 | down | yes |
| ENSG00000173276 | ZBTB21 | ENST00000310826;ENST00000425521;ENST00000398497 | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0008327(methyl-CpG binding);GO:0008327(methyl-CpG binding) | NA | NA | NA | zinc finger and BTB domain containing 21 [Source:HGNC Symbol;Acc:HGNC:13083] | 1.50 | 0.15 | 9.78 | 3.29 | 5.00e-06 | 0.00 | up | yes |
| ENSG00000273344 | PAXIP1-AS1 | ENST00000608317 | NA | NA | NA | NA | PAXIP1 antisense RNA 1 (head to head) [Source:HGNC Symbol;Acc:HGNC:27328] | 0.38 | 3.91 | 0.10 | -3.35003642938142 | 5.36e-06 | 0.00 | down | yes |
| ENSG00000172840 | PDP2 | ENST00000566543;ENST00000561704;ENST00000568398;ENST00000568720;ENST00000311765 | GO:0003824(catalytic activity);GO:0006470(protein dephosphorylation);GO:0004722(protein serine/threonine phosphatase activity);GO:0043169(cation binding);GO:0046872(metal ion binding);GO:0016787(hydrolase activity);GO:0005739(mitochondrion);GO:0004721(phosphoprotein phosphatase activity);GO:0005759(mitochondrial matrix);GO:0010510(regulation of acetyl-CoA biosynthetic process from pyruvate);GO:0004741([pyruvate dehydrogenase (lipoamide)] phosphatase activity);GO:0004741([pyruvate dehydrogenase (lipoamide)] phosphatase activity) | NA | NA | NA | pyruvate dehyrogenase phosphatase catalytic subunit 2 [Source:HGNC Symbol;Acc:HGNC:30263] | 0.10 | 1.03 | 0.10 | -3.35046626927724 | 5.59e-06 | 0.00 | down | yes |
| ENSG00000131446 | MGAT1 | ENST00000333055;ENST00000307826;ENST00000510341;ENST00000514760;ENST00000506708;ENST00000506033;ENST00000502678;ENST00000514283 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0046872(metal ion binding);GO:0016740(transferase activity);GO:0016757(transferase activity, transferring glycosyl groups);GO:0006486(protein glycosylation);GO:0005794(Golgi apparatus);GO:0000139(Golgi membrane);GO:0008375(acetylglucosaminyltransferase activity);GO:0070062(extracellular exosome);GO:0005975(carbohydrate metabolic process);GO:1903561(extracellular vesicle);GO:0003827(alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity);GO:0001701(in utero embryonic development);GO:0006049(UDP-N-acetylglucosamine catabolic process);GO:0006049(UDP-N-acetylglucosamine catabolic process) | 00510(N-Glycan biosynthesis);00510(N-Glycan biosynthesis) | K00726 | EC:2.4.1.101 | mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase [Source:HGNC Symbol;Acc:HGNC:7044] | 0.59 | 5.74 | 0.10 | -3.27549019731259 | 5.70e-06 | 0.00 | down | yes |
| ENSG00000265683 | SYPL1P2 | ENST00000582878 | NA | NA | NA | NA | synaptophysin like 1 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:53547] | 1.42 | 13.711954 | 0.10 | -3.27454185600457 | 7.71e-06 | 0.00 | down | yes |
| ENSG00000181924 | COA4 | ENST00000544575;ENST00000355693;ENST00000541455;ENST00000537581 | GO:0005739(mitochondrion);GO:0033617(mitochondrial respiratory chain complex IV assembly);GO:0005758(mitochondrial intermembrane space);GO:0005758(mitochondrial intermembrane space) | NA | NA | NA | cytochrome c oxidase assembly factor 4 homolog [Source:HGNC Symbol;Acc:HGNC:24604] | 20.800728371772 | 2.36 | 8.80 | 3.14 | 8.62e-06 | 0.00 | up | yes |
| ENSG00000277957 | SENP3-EIF4A1 | ENST00000614237 | GO:0008234(cysteine-type peptidase activity);GO:0006508(proteolysis);GO:0005634(nucleus);GO:0004175(endopeptidase activity);GO:0016929(SUMO-specific protease activity);GO:0016926(protein desumoylation) | NA | NA | NA | SENP3-EIF4A1 readthrough (NMD candidate) [Source:HGNC Symbol;Acc:HGNC:49182] | 0.21 | 2.03 | 0.10 | -3.26642216568654 | 9.36e-06 | 0.00 | down | yes |
| ENSG00000042429 | MED17 | ENST00000525613;ENST00000507258;ENST00000640411;ENST00000640077;ENST00000251871;ENST00000640027 | GO:0005634(nucleus);GO:0016020(membrane);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0001104(RNA polymerase II transcription cofactor activity);GO:0006357(regulation of transcription from RNA polymerase II promoter);GO:0016592(mediator complex);GO:0005515(protein binding);GO:0030374(ligand-dependent nuclear receptor transcription coactivator activity);GO:0045893(positive regulation of transcription, DNA-templated);GO:0006367(transcription initiation from RNA polymerase II promoter);GO:0005654(nucleoplasm);GO:0004872(receptor activity);GO:0003713(transcription coactivator activity);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0005667(transcription factor complex);GO:0003712(transcription cofactor activity);GO:0030518(intracellular steroid hormone receptor signaling pathway);GO:0030521(androgen receptor signaling pathway);GO:0042809(vitamin D receptor binding);GO:0046966(thyroid hormone receptor binding);GO:0046966(thyroid hormone receptor binding) | 04919(Thyroid hormone signaling pathway);04919(Thyroid hormone signaling pathway) | K15133 | NA | mediator complex subunit 17 [Source:HGNC Symbol;Acc:HGNC:2375] | 0.08 | 0.72 | 0.11 | -3.24592256759937 | 9.43e-06 | 0.00 | down | yes |
| ENSG00000264364 | DYNLL2 | ENST00000579991 | GO:0005739(mitochondrion);GO:0006810(transport);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0005874(microtubule);GO:0003774(motor activity);GO:0005813(centrosome);GO:0030286(dynein complex);GO:0005886(plasma membrane);GO:0007017(microtubule-based process);GO:0035735(intraciliary transport involved in cilium assembly);GO:0005929(cilium);GO:0097542(ciliary tip);GO:0005829(cytosol);GO:0060271(cilium assembly);GO:0051959(dynein light intermediate chain binding);GO:0007062(sister chromatid cohesion);GO:0019886(antigen processing and presentation of exogenous peptide antigen via MHC class II);GO:0006888(ER to Golgi vesicle-mediated transport);GO:0008574(ATP-dependent microtubule motor activity, plus-end-directed);GO:2000582(positive regulation of ATP-dependent microtubule motor activity, plus-end-directed);GO:0005868(cytoplasmic dynein complex);GO:1900740(positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway);GO:0016236(macroautophagy);GO:0008092(cytoskeletal protein binding);GO:0045505(dynein intermediate chain binding);GO:0010970(transport along microtubule);GO:0005875(microtubule associated complex);GO:0008039(synaptic target recognition);GO:0016459(myosin complex);GO:0031475(myosin V complex) | 04962(Vasopressin-regulated water reabsorption) | K10418 | NA | dynein light chain LC8-type 2 [Source:HGNC Symbol;Acc:HGNC:24596] | 9.73 | 1.13 | 8.61 | 3.11 | 9.79e-06 | 0.00 | up | yes |
| ENSG00000186814 | ZSCAN30 | ENST00000588832;ENST00000588448;ENST00000639929;ENST00000420878;ENST00000592598 | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0006355(regulation of transcription, DNA-templated);GO:0006351(transcription, DNA-templated);GO:0003677(DNA binding);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0003700(transcription factor activity, sequence-specific DNA binding) | NA | NA | NA | zinc finger and SCAN domain containing 30 [Source:HGNC Symbol;Acc:HGNC:33517] | 0.96 | 0.11 | 8.87 | 3.15 | 1.22e-05 | 0.00 | up | yes |
| ENSG00000158122 | AAED1 | ENST00000375234 | GO:0098869(cellular oxidant detoxification);GO:0055114(oxidation-reduction process);GO:0016209(antioxidant activity) | NA | NA | NA | AhpC/TSA antioxidant enzyme domain containing 1 [Source:HGNC Symbol;Acc:HGNC:16881] | 3.07 | 0.35 | 8.67 | 3.12 | 1.26e-05 | 0.00 | up | yes |
| ENSG00000171310 | CHST11 | ENST00000550711;ENST00000549016;ENST00000547956;ENST00000303694 | GO:0016021(integral component of membrane);GO:0016740(transferase activity);GO:0005794(Golgi apparatus);GO:0000139(Golgi membrane);GO:0008146(sulfotransferase activity);GO:0005975(carbohydrate metabolic process);GO:0016051(carbohydrate biosynthetic process);GO:0016020(membrane);GO:0030206(chondroitin sulfate biosynthetic process);GO:0001537(N-acetylgalactosamine 4-O-sulfotransferase activity);GO:0047756(chondroitin 4-sulfotransferase activity);GO:0050659(N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity);GO:0002063(chondrocyte development);GO:0007585(respiratory gaseous exchange);GO:0009791(post-embryonic development);GO:0030204(chondroitin sulfate metabolic process);GO:0030326(embryonic limb morphogenesis);GO:0030512(negative regulation of transforming growth factor beta receptor signaling pathway);GO:0033037(polysaccharide localization);GO:0036342(post-anal tail morphogenesis);GO:0042127(regulation of cell proliferation);GO:0042733(embryonic digit morphogenesis);GO:0043066(negative regulation of apoptotic process);GO:0048589(developmental growth);GO:0048703(embryonic viscerocranium morphogenesis);GO:0048704(embryonic skeletal system morphogenesis);GO:0051216(cartilage development);GO:0051216(cartilage development) | 00532(Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate);00532(Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate) | K01017 | EC:2.8.2.5 | carbohydrate sulfotransferase 11 [Source:HGNC Symbol;Acc:HGNC:17422] | 0.77 | 6.67 | 0.11 | -3.12129884162869 | 1.35e-05 | 0.00 | down | yes |
| ENSG00000168818 | STX18 | ENST00000505286;ENST00000515687;ENST00000503692 | GO:0005622(intracellular);GO:0006886(intracellular protein transport);GO:0016020(membrane);GO:0005484(SNAP receptor activity);GO:0061025(membrane fusion);GO:0006810(transport);GO:0016021(integral component of membrane);GO:0015031(protein transport);GO:0005794(Golgi apparatus);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0005515(protein binding);GO:0000139(Golgi membrane);GO:0016192(vesicle-mediated transport);GO:0006890(retrograde vesicle-mediated transport, Golgi to ER);GO:0031201(SNARE complex);GO:1903358(regulation of Golgi organization);GO:1902953(positive regulation of ER to Golgi vesicle-mediated transport);GO:1902117(positive regulation of organelle assembly);GO:0019904(protein domain specific binding);GO:0090158(endoplasmic reticulum membrane organization);GO:0090158(endoplasmic reticulum membrane organization) | 04130(SNARE interactions in vesicular transport);04145(Phagosome);04145(Phagosome) | NA | NA | syntaxin 18 [Source:HGNC Symbol;Acc:HGNC:15942] | 2.07 | 0.25 | 8.44 | 3.08 | 1.47e-05 | 0.00 | up | yes |
| ENSG00000143570 | SLC39A1 | ENST00000368623;ENST00000621013;ENST00000310483 | GO:0006810(transport);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0055085(transmembrane transport);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0006811(ion transport);GO:0005886(plasma membrane);GO:0046873(metal ion transmembrane transporter activity);GO:0030001(metal ion transport);GO:0005385(zinc ion transmembrane transporter activity);GO:0006829(zinc II ion transport);GO:0071577(zinc II ion transmembrane transport);GO:0022890(inorganic cation transmembrane transporter activity);GO:0006812(cation transport);GO:0005102(receptor binding);GO:0001701(in utero embryonic development);GO:0048701(embryonic cranial skeleton morphogenesis);GO:0060173(limb development);GO:0060173(limb development) | NA | NA | NA | solute carrier family 39 member 1 [Source:HGNC Symbol;Acc:HGNC:12876] | 89.8272316623377 | 11.0805413719008 | 8.11 | 3.02 | 1.52e-05 | 0.00 | up | yes |
| ENSG00000213516 | RBMXL1 | ENST00000399794 | GO:0003676(nucleic acid binding);GO:0003723(RNA binding);GO:0006397(mRNA processing);GO:0008380(RNA splicing);GO:0005634(nucleus);GO:0030529(intracellular ribonucleoprotein complex);GO:0001047(core promoter binding);GO:0003682(chromatin binding);GO:0045944(positive regulation of transcription from RNA polymerase II promoter) | 03040(Spliceosome) | K12885 | NA | RNA binding motif protein, X-linked like 1 [Source:HGNC Symbol;Acc:HGNC:25073] | 18.9055061151956 | 2.33 | 8.11 | 3.02 | 1.55e-05 | 0.00 | up | yes |
| ENSG00000166881 | NEMP1 | ENST00000379391;ENST00000300128;ENST00000553654 | GO:0005634(nucleus);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0003674(molecular_function);GO:0008150(biological_process);GO:0005637(nuclear inner membrane);GO:0005635(nuclear envelope);GO:0005635(nuclear envelope) | NA | NA | NA | nuclear envelope integral membrane protein 1 [Source:HGNC Symbol;Acc:HGNC:29001] | 10.9388675315347 | 1.35 | 8.12 | 3.02 | 1.60e-05 | 0.00 | up | yes |
| ENSG00000101220 | C20orf27 | ENST00000379772;ENST00000217195 | GO:0003674(molecular_function);GO:0008150(biological_process);GO:0005575(cellular_component);GO:0005575(cellular_component) | NA | NA | NA | chromosome 20 open reading frame 27 [Source:HGNC Symbol;Acc:HGNC:15873] | 19.4631206857143 | 2.40 | 8.11 | 3.02 | 1.62e-05 | 0.00 | up | yes |
| ENSG00000186470 | BTN3A2 | ENST00000508906;ENST00000604202 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005886(plasma membrane);GO:0002376(immune system process);GO:0002250(adaptive immune response);GO:0002456(T cell mediated immunity);GO:0072643(interferon-gamma secretion);GO:0072643(interferon-gamma secretion) | NA | NA | NA | butyrophilin subfamily 3 member A2 [Source:HGNC Symbol;Acc:HGNC:1139] | 0.14 | 1.19 | 0.11 | -3.13752324056438 | 1.73e-05 | 0.00 | down | yes |
| ENSG00000259399 | TGIF2-C20orf24 | ENST00000558530 | GO:0005634(nucleus);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0005739(mitochondrion) | NA | NA | NA | TGIF2-C20orf24 readthrough [Source:HGNC Symbol;Acc:HGNC:44664] | 38.181866 | 4.81 | 7.94 | 2.99 | 1.97e-05 | 0.00 | up | yes |
| ENSG00000105647 | PIK3R2 | ENST00000222254;ENST00000600533;ENST00000426902;ENST00000459743 | GO:0043409(negative regulation of MAPK cascade);GO:0006810(transport);GO:0005634(nucleus);GO:0015031(protein transport);GO:0007165(signal transduction);GO:0043547(positive regulation of GTPase activity);GO:0005515(protein binding);GO:0036092(phosphatidylinositol-3-phosphate biosynthetic process);GO:0038095(Fc-epsilon receptor signaling pathway);GO:0038096(Fc-gamma receptor signaling pathway involved in phagocytosis);GO:0050900(leukocyte migration);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0005829(cytosol);GO:0005096(GTPase activator activity);GO:0051056(regulation of small GTPase mediated signal transduction);GO:0050852(T cell receptor signaling pathway);GO:0035014(phosphatidylinositol 3-kinase regulator activity);GO:0005942(phosphatidylinositol 3-kinase complex);GO:0046854(phosphatidylinositol phosphorylation);GO:0046935(1-phosphatidylinositol-3-kinase regulator activity);GO:0008286(insulin receptor signaling pathway);GO:0043551(regulation of phosphatidylinositol 3-kinase activity);GO:2001275(positive regulation of glucose import in response to insulin stimulus);GO:0019903(protein phosphatase binding);GO:0046982(protein heterodimerization activity);GO:0046934(phosphatidylinositol-4,5-bisphosphate 3-kinase activity);GO:0006661(phosphatidylinositol biosynthetic process);GO:0014065(phosphatidylinositol 3-kinase signaling);GO:0014066(regulation of phosphatidylinositol 3-kinase signaling);GO:0038111(interleukin-7-mediated signaling pathway);GO:0048015(phosphatidylinositol-mediated signaling);GO:0001784(phosphotyrosine residue binding);GO:0016303(1-phosphatidylinositol-3-kinase activity);GO:0030971(receptor tyrosine kinase binding);GO:0001678(cellular glucose homeostasis);GO:0010506(regulation of autophagy);GO:0032869(cellular response to insulin stimulus);GO:0034976(response to endoplasmic reticulum stress);GO:0042993(positive regulation of transcription factor import into nucleus);GO:0048010(vascular endothelial growth factor receptor signaling pathway);GO:0048010(vascular endothelial growth factor receptor signaling pathway) | 04014(Ras signaling pathway);04015(Rap1 signaling pathway);04012(ErbB signaling pathway);04370(VEGF signaling pathway);04630(Jak-STAT signaling pathway);04668(TNF signaling pathway);04066(HIF-1 signaling pathway);04068(FoxO signaling pathway);04070(Phosphatidylinositol signaling system);04072(Phospholipase D signaling pathway);04071(Sphingolipid signaling pathway);04024(cAMP signaling pathway);04151(PI3K-Akt signaling pathway);04152(AMPK signaling pathway);04150(mTOR signaling pathway);04140(Autophagy - animal);04210(Apoptosis);04218(Cellular senescence);04510(Focal adhesion);04550(Signaling pathways regulating pluripotency of stem cells);04810(Regulation of actin cytoskeleton);04611(Platelet activation);04620(Toll-like receptor signaling pathway);04650(Natural killer cell mediated cytotoxicity);04660(T cell receptor signaling pathway);04662(B cell receptor signaling pathway);04664(Fc epsilon RI signaling pathway);04666(Fc gamma R-mediated phagocytosis);04670(Leukocyte transendothelial migration);04062(Chemokine signaling pathway);04910(Insulin signaling pathway);04923(Regulation of lipolysis in adipocyte);04915(Estrogen signaling pathway);04914(Progesterone-mediated oocyte maturation);04917(Prolactin signaling pathway);04926(Relaxin signaling pathway);04919(Thyroid hormone signaling pathway);04973(Carbohydrate digestion and absorption);04960(Aldosterone-regulated sodium reabsorption);04725(Cholinergic synapse);04722(Neurotrophin signaling pathway);04750(Inflammatory mediator regulation of TRP channels);04360(Axon guidance);04380(Osteoclast differentiation);04211(Longevity regulating pathway - mammal);04213(Longevity regulating pathway - multiple species);05200(Pathways in cancer);05230(Central carbon metabolism in cancer);05231(Choline metabolism in cancer);05206(MicroRNAs in cancer);05205(Proteoglycans in cancer);05203(Viral carcinogenesis);05210(Colorectal cancer);05212(Pancreatic cancer);05225(Hepatocellular carcinoma);05226(Gastric cancer);05214(Glioma);05221(Acute myeloid leukemia);05220(Chronic myeloid leukemia);05218(Melanoma);05211(Renal cell carcinoma);05215(Prostate cancer);05213(Endometrial cancer);05224(Breast cancer);05222(Small cell lung cancer);05223(Non-small cell lung cancer);05418(Fluid shear stress and atherosclerosis);04930(Type II diabetes mellitus);04932(Non-alcoholic fatty liver disease (NAFLD));04931(Insulin resistance);04933(AGE-RAGE signaling pathway in diabetic complications);05100(Bacterial invasion of epithelial cells);05166(HTLV-I infection);05162(Measles);05164(Influenza A);05161(Hepatitis B);05160(Hepatitis C);05167(Kaposi's sarcoma-associated herpesvirus infection);05169(Epstein-Barr virus infection);05165(Human papillomavirus infection);05146(Amoebiasis);05142(Chagas disease (American trypanosomiasis));01521(EGFR tyrosine kinase inhibitor resistance);01524(Platinum drug resistance);01522(Endocrine resistance);01522(Endocrine resistance) | K02649 | NA | phosphoinositide-3-kinase regulatory subunit 2 [Source:HGNC Symbol;Acc:HGNC:8980] | 20.50887958687 | 2.64 | 7.78 | 2.96 | 2.16e-05 | 0.00 | up | yes |
| ENSG00000261408 | TEN1-CDK3 | ENST00000567351 | NA | NA | NA | NA | TEN1-CDK3 readthrough (NMD candidate) [Source:HGNC Symbol;Acc:HGNC:44420] | 0.26 | 2.19 | 0.12 | -3.08647331256259 | 2.36e-05 | 0.00 | down | yes |
| ENSG00000180329 | CCDC43 | ENST00000588210;ENST00000457422;ENST00000315286;ENST00000588687 | GO:0005515(protein binding);GO:0005515(protein binding) | NA | NA | NA | coiled-coil domain containing 43 [Source:HGNC Symbol;Acc:HGNC:26472] | 0.97 | 7.90 | 0.12 | -3.03330437652278 | 2.49e-05 | 0.00 | down | yes |
| ENSG00000175548 | ALG10B | ENST00000553138;ENST00000308742 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0016740(transferase activity);GO:0016757(transferase activity, transferring glycosyl groups);GO:0005783(endoplasmic reticulum);GO:0005886(plasma membrane);GO:0005789(endoplasmic reticulum membrane);GO:0006488(dolichol-linked oligosaccharide biosynthetic process);GO:0006486(protein glycosylation);GO:0004583(dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity);GO:0060050(positive regulation of protein glycosylation);GO:1901980(positive regulation of inward rectifier potassium channel activity);GO:1901980(positive regulation of inward rectifier potassium channel activity) | 00510(N-Glycan biosynthesis);00510(N-Glycan biosynthesis) | K03850 | EC:2.4.1.256 | ALG10B, alpha-1,2-glucosyltransferase [Source:HGNC Symbol;Acc:HGNC:31088] | 0.10 | 0.84 | 0.12 | -3.02466346231686 | 2.81e-05 | 0.00 | down | yes |
| ENSG00000168283 | BMI1 | ENST00000376663;ENST00000496174;ENST00000602524;ENST00000456675;ENST00000602358;ENST00000442508 | GO:0048146(positive regulation of fibroblast proliferation);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0005829(cytosol);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0016569(covalent chromatin modification);GO:0008270(zinc ion binding);GO:0005515(protein binding);GO:0016604(nuclear body);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0005654(nucleoplasm);GO:0031519(PcG protein complex);GO:0035102(PRC1 complex);GO:0030097(hemopoiesis);GO:0000151(ubiquitin ligase complex);GO:1990841(promoter-specific chromatin binding);GO:0045814(negative regulation of gene expression, epigenetic);GO:0010468(regulation of gene expression);GO:0071535(RING-like zinc finger domain binding);GO:0007379(segment specification);GO:0051443(positive regulation of ubiquitin-protein transferase activity);GO:0051443(positive regulation of ubiquitin-protein transferase activity) | 04550(Signaling pathways regulating pluripotency of stem cells);05202(Transcriptional misregulation in cancers);05206(MicroRNAs in cancer);05206(MicroRNAs in cancer) | K11459 | NA | BMI1 proto-oncogene, polycomb ring finger [Source:HGNC Symbol;Acc:HGNC:1066] | 7.17 | 0.95 | 7.58 | 2.92 | 2.88e-05 | 0.00 | up | yes |
| ENSG00000229807 | XIST | ENST00000434839;ENST00000602587;ENST00000429829;ENST00000635841;ENST00000602495 | NA | NA | NA | NA | X inactive specific transcript (non-protein coding) [Source:HGNC Symbol;Acc:HGNC:12810] | 0.09 | 0.71 | 0.12 | -3.00967201332762 | 3.06e-05 | 0.00 | down | yes |
| ENSG00000102100 | SLC35A2 | ENST00000376521;ENST00000635015 | GO:0006810(transport);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005794(Golgi apparatus);GO:0005634(nucleus);GO:0000139(Golgi membrane);GO:0008643(carbohydrate transport);GO:0005351(sugar:proton symporter activity);GO:0015992(proton transport);GO:0005783(endoplasmic reticulum);GO:0006012(galactose metabolic process);GO:0005459(UDP-galactose transmembrane transporter activity);GO:0015785(UDP-galactose transport);GO:0072334(UDP-galactose transmembrane transport);GO:0072334(UDP-galactose transmembrane transport) | NA | NA | NA | solute carrier family 35 member A2 [Source:HGNC Symbol;Acc:HGNC:11022] | 0.83 | 6.53 | 0.13 | -2.98536553691795 | 3.07e-05 | 0.00 | down | yes |
| ENSG00000178878 | APOLD1 | ENST00000540583;ENST00000326765;ENST00000534843 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0005576(extracellular region);GO:0005886(plasma membrane);GO:0042157(lipoprotein metabolic process);GO:0006869(lipid transport);GO:0008289(lipid binding);GO:0007275(multicellular organism development);GO:0030154(cell differentiation);GO:0001525(angiogenesis);GO:0001666(response to hypoxia);GO:0042118(endothelial cell activation);GO:0045601(regulation of endothelial cell differentiation);GO:0045601(regulation of endothelial cell differentiation) | NA | NA | NA | apolipoprotein L domain containing 1 [Source:HGNC Symbol;Acc:HGNC:25268] | 5.24 | 0.70 | 7.49 | 2.91 | 3.08e-05 | 0.00 | up | yes |
| ENSG00000162971 | TYW5 | ENST00000354611;ENST00000493181 | GO:0046872(metal ion binding);GO:0008033(tRNA processing);GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0042803(protein homodimerization activity);GO:0051213(dioxygenase activity);GO:0000049(tRNA binding);GO:0005506(iron ion binding);GO:0016706(oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors);GO:0006400(tRNA modification);GO:0031591(wybutosine biosynthetic process);GO:0005737(cytoplasm);GO:0005737(cytoplasm) | NA | NA | NA | tRNA-yW synthesizing protein 5 [Source:HGNC Symbol;Acc:HGNC:26754] | 0.55 | 0.07 | 7.87 | 2.98 | 3.52e-05 | 0.01 | up | yes |
Definition of table:
| term | 意义 |
|---|---|
| gene_id | The ID of gene, some ids started with "MSTRG" are named by transcript assembly software StringTie |
| gene_name | The name of gene (gene symbol) |
| transcript_id | The name of transcript, separated by comma |
| GO | Gene ontology term and description http://www.geneontology.org/ |
| KEGG | KEGG pathway annotation and description http://www.kegg.jp/kegg/kegg3a.html |
| KO_Entry | KO functional ortholog, KO groups identified by K numbers and, in metabolic maps |
| EC | enzyme |
| Description | Description of gene |
| FPKM.A | Normalization of gene expression level by FPKM value |
| fc | fold change |
| log2(fc) | log2 of fold change value |
| pval | p value |
| qval | q value or FDR value |
| regulation | up-regulation or down-regulation |
| significant | yes or no |
document location: summary/7_differential_expression/*VS*/*_Gene_differential_expression.xlsx
| t_id | class_code | chr | strand | start | end | t_name | num_exons | length | gene_id | gene_name | GO | KEGG | KO_ENTRY | EC | Description | FPKM.M14_OE | FPKM.M14_NC | fc | log2(fc) | pval | qval | regulation | significant |
| 40732 | NA | chr12 | - | 49127782 | 49131395 | ENST00000336023 | 4 | 1627 | ENSG00000123416 | TUBA1B | GO:0005737(cytoplasm);GO:0005525(GTP binding);GO:0000166(nucleotide binding);GO:0003924(GTPase activity);GO:0015630(microtubule cytoskeleton);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0005200(structural constituent of cytoskeleton);GO:0005874(microtubule);GO:0007017(microtubule-based process);GO:0070062(extracellular exosome);GO:0031625(ubiquitin protein ligase binding);GO:0003725(double-stranded RNA binding);GO:0005198(structural molecule activity);GO:0030705(cytoskeleton-dependent intracellular transport);GO:0051301(cell division);GO:0000226(microtubule cytoskeleton organization);GO:0071353(cellular response to interleukin-4);GO:0005881(cytoplasmic microtubule);GO:0043209(myelin sheath);GO:0045121(membrane raft);GO:0007010(cytoskeleton organization) | 04145(Phagosome);04210(Apoptosis);04530(Tight junction);04540(Gap junction);05130(Pathogenic Escherichia coli infection) | K07374 | NA | tubulin alpha 1b [Source:HGNC Symbol;Acc:HGNC:18809] | 0.61 | 1269.210693 | 0.00 | -11.0202661704784 | 2.61e-28 | 4.77e-24 | down | yes |
| 186296 | NA | chr8 | - | 143915153 | 143950876 | ENST00000322810 | 32 | 15249 | ENSG00000178209 | PLEC | GO:0003723(RNA binding);GO:0005737(cytoplasm);GO:0005829(cytosol);GO:0003779(actin binding);GO:0005925(focal adhesion);GO:0045111(intermediate filament cytoskeleton);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0008092(cytoskeletal protein binding);GO:0030054(cell junction);GO:0045296(cadherin binding);GO:0070062(extracellular exosome);GO:0042383(sarcolemma);GO:0031012(extracellular matrix);GO:0005886(plasma membrane);GO:0008307(structural constituent of muscle);GO:0030506(ankyrin binding);GO:0031581(hemidesmosome assembly);GO:0030056(hemidesmosome);GO:0043034(costamere);GO:0016528(sarcoplasm);GO:0005903(brush border);GO:0043292(contractile fiber) | NA | NA | NA | plectin [Source:HGNC Symbol;Acc:HGNC:9069] | 0.12 | 65.769226 | 0.00 | -9.15135392771124 | 8.91e-23 | 8.14e-19 | down | yes |
| 132693 | NA | chr3 | + | 22381945 | 22382929 | ENST00000451497 | 1 | 985 | ENSG00000132967 | HMGB1P5 | NA | NA | NA | NA | high mobility group box 1 pseudogene 5 [Source:HGNC Symbol;Acc:HGNC:4997] | 0.93 | 314.333008 | 0.00 | -8.39980990887748 | 2.08e-20 | 1.26e-16 | down | yes |
| 11017 | NA | chr1 | - | 153606886 | 153609337 | ENST00000368704 | 3 | 1146 | ENSG00000188643 | S100A16 | GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0005829(cytosol);GO:0005509(calcium ion binding);GO:0005886(plasma membrane);GO:0042803(protein homodimerization activity);GO:0005515(protein binding);GO:0005615(extracellular space);GO:0005730(nucleolus);GO:0070062(extracellular exosome);GO:0051592(response to calcium ion) | NA | NA | NA | S100 calcium binding protein A16 [Source:HGNC Symbol;Acc:HGNC:20441] | 355.696381 | 1.23 | 288.235216753954 | 8.17 | 5.06e-20 | 2.31e-16 | up | yes |
| 186292 | NA | chr8 | - | 143915147 | 143939590 | ENST00000345136 | 32 | 14803 | ENSG00000178209 | PLEC | GO:0003723(RNA binding);GO:0005737(cytoplasm);GO:0005829(cytosol);GO:0003779(actin binding);GO:0005925(focal adhesion);GO:0045111(intermediate filament cytoskeleton);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0008092(cytoskeletal protein binding);GO:0030054(cell junction);GO:0045296(cadherin binding);GO:0070062(extracellular exosome);GO:0042383(sarcolemma);GO:0031012(extracellular matrix);GO:0005886(plasma membrane);GO:0008307(structural constituent of muscle);GO:0030506(ankyrin binding);GO:0031581(hemidesmosome assembly);GO:0030056(hemidesmosome);GO:0043034(costamere);GO:0016528(sarcoplasm);GO:0005903(brush border);GO:0043292(contractile fiber) | NA | NA | NA | plectin [Source:HGNC Symbol;Acc:HGNC:9069] | 67.527359 | 0.31 | 220.567360110794 | 7.79 | 5.57e-19 | 2.03e-15 | up | yes |
| 113163 | NA | chr2 | - | 130151392 | 130181757 | ENST00000431183 | 17 | 3614 | ENSG00000136699 | SMPD4 | GO:0050290(sphingomyelin phosphodiesterase D activity);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0071356(cellular response to tumor necrosis factor);GO:0046872(metal ion binding);GO:0016787(hydrolase activity);GO:0005794(Golgi apparatus);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0000139(Golgi membrane);GO:0006687(glycosphingolipid metabolic process);GO:0004767(sphingomyelin phosphodiesterase activity);GO:0046513(ceramide biosynthetic process);GO:0046475(glycerophospholipid catabolic process);GO:0006685(sphingomyelin catabolic process);GO:0005802(trans-Golgi network) | 00600(Sphingolipid metabolism) | K12353 | EC:3.1.4.12 | sphingomyelin phosphodiesterase 4 [Source:HGNC Symbol;Acc:HGNC:32949] | 31.924562 | 0.19 | 164.773633792349 | 7.36 | 1.99e-17 | 6.07e-14 | up | yes |
| 152173 | NA | chr5 | - | 17353695 | 17354624 | ENST00000401830 | 1 | 930 | ENSG00000223361 | FTH1P10 | NA | NA | NA | NA | ferritin heavy chain 1 pseudogene 10 [Source:HGNC Symbol;Acc:HGNC:3980] | 1.64 | 251.200851 | 0.01 | -7.25830254086227 | 4.20e-17 | 1.09e-13 | down | yes |
| 135852 | NA | chr3 | - | 53225652 | 53235607 | ENST00000460343 | 6 | 7286 | ENSG00000163931 | TKT | GO:0016607(nuclear speck);GO:0046872(metal ion binding);GO:0005654(nucleoplasm);GO:0016740(transferase activity);GO:0003824(catalytic activity);GO:0008152(metabolic process);GO:0042803(protein homodimerization activity);GO:0005515(protein binding);GO:0048037(cofactor binding);GO:0016604(nuclear body);GO:0005777(peroxisome);GO:0005829(cytosol);GO:0070062(extracellular exosome);GO:0031982(vesicle);GO:0006098(pentose-phosphate shunt);GO:0004802(transketolase activity);GO:0005999(xylulose biosynthetic process);GO:0009052(pentose-phosphate shunt, non-oxidative branch);GO:0046166(glyceraldehyde-3-phosphate biosynthetic process);GO:0040008(regulation of growth);GO:0043209(myelin sheath) | 00030(Pentose phosphate pathway);01051(Biosynthesis of ansamycins) | NA | NA | transketolase [Source:HGNC Symbol;Acc:HGNC:11834] | 0.20 | 28.908144 | 0.01 | -7.17326339230639 | 7.75e-17 | 1.76e-13 | down | yes |
| 41959 | NA | chr12 | + | 54325127 | 54351849 | ENST00000262061 | 9 | 1897 | ENSG00000111481 | COPZ1 | GO:0006810(transport);GO:0006886(intracellular protein transport);GO:0015031(protein transport);GO:0016192(vesicle-mediated transport);GO:0030117(membrane coat);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0005794(Golgi apparatus);GO:0000139(Golgi membrane);GO:0031410(cytoplasmic vesicle);GO:0030663(COPI-coated vesicle membrane);GO:0005789(endoplasmic reticulum membrane);GO:0005829(cytosol);GO:0006890(retrograde vesicle-mediated transport, Golgi to ER);GO:0006888(ER to Golgi vesicle-mediated transport);GO:0030133(transport vesicle);GO:0006891(intra-Golgi vesicle-mediated transport);GO:0030126(COPI vesicle coat);GO:1901998(toxin transport) | NA | NA | NA | coatomer protein complex subunit zeta 1 [Source:HGNC Symbol;Acc:HGNC:2243] | 0.64 | 81.771904 | 0.01 | -7.00028231431718 | 2.50e-16 | 4.70e-13 | down | yes |
| 148905 | NA | chr4 | + | 118685368 | 118715433 | ENST00000388822 | 11 | 6669 | ENSG00000145388 | METTL14 | GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005654(nucleoplasm);GO:0008168(methyltransferase activity);GO:0016740(transferase activity);GO:0032259(methylation);GO:0006139(nucleobase-containing compound metabolic process);GO:0003729(mRNA binding);GO:0005515(protein binding);GO:0000398(mRNA splicing, via spliceosome);GO:0016070(RNA metabolic process);GO:0019827(stem cell population maintenance);GO:0061157(mRNA destabilization);GO:0080009(mRNA methylation);GO:0036396(MIS complex);GO:0016422(mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity) | NA | NA | NA | methyltransferase like 14 [Source:HGNC Symbol;Acc:HGNC:29330] | 116.351814 | 1.00 | 116.925215255614 | 6.87 | 2.82e-16 | 4.70e-13 | up | yes |
| 96594 | NA | chr19 | - | 14481747 | 14483463 | ENST00000587811 | 1 | 1717 | ENSG00000123159 | GIPC1 | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0005829(cytosol);GO:0005515(protein binding);GO:0007186(G-protein coupled receptor signaling pathway);GO:0031410(cytoplasmic vesicle);GO:0003779(actin binding);GO:0017022(myosin binding);GO:0030165(PDZ domain binding);GO:0042803(protein homodimerization activity);GO:0006605(protein targeting);GO:0007268(chemical synaptic transmission);GO:0014047(glutamate secretion);GO:0030511(positive regulation of transforming growth factor beta receptor signaling pathway);GO:0031647(regulation of protein stability);GO:0032435(negative regulation of proteasomal ubiquitin-dependent protein catabolic process);GO:0043542(endothelial cell migration);GO:0048167(regulation of synaptic plasticity);GO:0005903(brush border);GO:0008021(synaptic vesicle);GO:0012506(vesicle membrane);GO:0030139(endocytic vesicle);GO:0043197(dendritic spine);GO:0043198(dendritic shaft);GO:0045296(cadherin binding);GO:0070062(extracellular exosome);GO:0005938(cell cortex);GO:0005102(receptor binding);GO:0032467(positive regulation of cytokinesis) | NA | NA | NA | GIPC PDZ domain containing family member 1 [Source:HGNC Symbol;Acc:HGNC:1226] | 43.676731 | 0.35 | 126.156028676071 | 6.98 | 2.83e-16 | 4.70e-13 | up | yes |
| 97483 | NA | chr19 | + | 18153158 | 18178117 | ENST00000593731 | 25 | 5142 | ENSG00000268173 | AC007192.1 | NA | NA | NA | NA | NA | 0.13 | 15.335408 | 0.01 | -6.83520972451006 | 1.07e-15 | 1.51e-12 | down | yes |
| 42528 | NA | chr12 | - | 56271699 | 56300318 | ENST00000549143 | 11 | 2811 | ENSG00000062485 | CS | GO:0016740(transferase activity);GO:0006099(tricarboxylic acid cycle);GO:0005739(mitochondrion);GO:0046912(transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer);GO:0006101(citrate metabolic process);GO:0004108(citrate (Si)-synthase activity);GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005759(mitochondrial matrix);GO:0005975(carbohydrate metabolic process);GO:0070062(extracellular exosome);GO:0016020(membrane);GO:0016021(integral component of membrane) | 00020(Citrate cycle (TCA cycle));00630(Glyoxylate and dicarboxylate metabolism) | NA | NA | citrate synthase [Source:HGNC Symbol;Acc:HGNC:2422] | 44.738632 | 0.42 | 106.738604100758 | 6.74 | 1.08e-15 | 1.51e-12 | up | yes |
| 92969 | NA | chr19 | + | 572565 | 583490 | ENST00000333511 | 9 | 1974 | ENSG00000172270 | BSG | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0043231(intracellular membrane-bounded organelle);GO:0030246(carbohydrate binding);GO:0005739(mitochondrion);GO:0005886(plasma membrane);GO:0042470(melanosome);GO:0005925(focal adhesion);GO:0050900(leukocyte migration);GO:0005887(integral component of plasma membrane);GO:0005515(protein binding);GO:0045296(cadherin binding);GO:0070062(extracellular exosome);GO:0007166(cell surface receptor signaling pathway);GO:0006090(pyruvate metabolic process);GO:0000139(Golgi membrane);GO:0030198(extracellular matrix organization);GO:0022617(extracellular matrix disassembly);GO:0008028(monocarboxylic acid transmembrane transporter activity);GO:0005537(mannose binding);GO:0015718(monocarboxylic acid transport);GO:0072661(protein targeting to plasma membrane);GO:0007566(embryo implantation);GO:0042475(odontogenesis of dentin-containing tooth);GO:0043434(response to peptide hormone);GO:0046689(response to mercury ion);GO:0046697(decidualization);GO:0051591(response to cAMP);GO:0002080(acrosomal membrane);GO:0042383(sarcolemma);GO:0045121(membrane raft) | NA | NA | NA | basigin (Ok blood group) [Source:HGNC Symbol;Acc:HGNC:1116] | 1.32 | 134.203247 | 0.01 | -6.66937929022245 | 1.97e-15 | 2.57e-12 | down | yes |
| 31382 | NA | chr11 | + | 66616631 | 66644632 | ENST00000421355 | 3 | 2034 | ENSG00000248643 | RBM14-RBM4 | GO:0003676(nucleic acid binding);GO:0003723(RNA binding) | NA | NA | NA | RBM14-RBM4 readthrough [Source:HGNC Symbol;Acc:HGNC:38840] | 28.006208 | 0.27 | 101.880026482935 | 6.67 | 2.33e-15 | 2.84e-12 | up | yes |
| 175804 | NA | chr7 | - | 106090503 | 106112331 | ENST00000455385 | 5 | 2151 | ENSG00000008282 | SYPL1 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0006810(transport);GO:0005215(transporter activity);GO:0008021(synaptic vesicle);GO:0042470(melanosome);GO:0030659(cytoplasmic vesicle membrane);GO:0031410(cytoplasmic vesicle);GO:0070062(extracellular exosome);GO:0005887(integral component of plasma membrane);GO:0007268(chemical synaptic transmission);GO:0030285(integral component of synaptic vesicle membrane);GO:0030141(secretory granule) | NA | NA | NA | synaptophysin like 1 [Source:HGNC Symbol;Acc:HGNC:11507] | 0.31 | 31.097704 | 0.01 | -6.65601119576331 | 3.71e-15 | 4.24e-12 | down | yes |
| 121376 | NA | chr20 | - | 25300198 | 25390976 | ENST00000339157 | 13 | 2117 | ENSG00000100997 | ABHD12 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0016787(hydrolase activity);GO:0005886(plasma membrane);GO:0047372(acylglycerol lipase activity);GO:0052651(monoacylglycerol catabolic process);GO:0046464(acylglycerol catabolic process);GO:0004622(lysophospholipase activity);GO:0006660(phosphatidylserine catabolic process);GO:0007628(adult walking behavior);GO:0010996(response to auditory stimulus);GO:0046475(glycerophospholipid catabolic process);GO:0032281(AMPA glutamate receptor complex) | NA | NA | NA | abhydrolase domain containing 12 [Source:HGNC Symbol;Acc:HGNC:15868] | 0.90 | 81.812836 | 0.01 | -6.50426245064507 | 6.58e-15 | 7.06e-12 | down | yes |
| 172305 | NA | chr7 | - | 50590068 | 50792969 | ENST00000403097 | 18 | 5432 | ENSG00000106070 | GRB10 | GO:0005737(cytoplasm);GO:0007165(signal transduction);GO:0005515(protein binding);GO:0005158(insulin receptor binding);GO:0005070(SH3/SH2 adaptor activity);GO:0005829(cytosol);GO:0030178(negative regulation of Wnt signaling pathway);GO:0043234(protein complex);GO:0046627(negative regulation of insulin receptor signaling pathway);GO:0005886(plasma membrane);GO:0007411(axon guidance);GO:0030949(positive regulation of vascular endothelial growth factor receptor signaling pathway);GO:0046325(negative regulation of glucose import);GO:0009967(positive regulation of signal transduction);GO:0042326(negative regulation of phosphorylation);GO:0048009(insulin-like growth factor receptor signaling pathway) | 04150(mTOR signaling pathway) | K20064 | NA | growth factor receptor bound protein 10 [Source:HGNC Symbol;Acc:HGNC:4564] | 17.421515 | 0.21 | 82.750748111908 | 6.37 | 1.29e-14 | 1.31e-11 | up | yes |
| 79058 | NA | chr17 | + | 16439327 | 16441779 | ENST00000491009 | 3 | 863 | ENSG00000175061 | LRRC75A-AS1 | NA | NA | NA | NA | LRRC75A antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:28619] | 313.477753 | 4.20 | 74.6819426497271 | 6.22 | 2.49e-14 | 2.39e-11 | up | yes |
| 8702 | NA | chr1 | - | 109309568 | 109393357 | ENST00000538502 | 20 | 6993 | ENSG00000134243 | SORT1 | GO:0048471(perinuclear region of cytoplasm);GO:0051005(negative regulation of lipoprotein lipase activity);GO:0006810(transport);GO:0005634(nucleus);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005794(Golgi apparatus);GO:0005829(cytosol);GO:0005765(lysosomal membrane);GO:0005886(plasma membrane);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0005515(protein binding);GO:0007275(multicellular organism development);GO:0030154(cell differentiation);GO:0005764(lysosome);GO:0032580(Golgi cisterna membrane);GO:0005768(endosome);GO:0006897(endocytosis);GO:0010008(endosome membrane);GO:0001503(ossification);GO:0009986(cell surface);GO:0031965(nuclear membrane);GO:0016050(vesicle organization);GO:0031410(cytoplasmic vesicle);GO:0019899(enzyme binding);GO:0005769(early endosome);GO:0007186(G-protein coupled receptor signaling pathway);GO:0005905(clathrin-coated pit);GO:0008625(extrinsic apoptotic signaling pathway via death domain receptors);GO:0007218(neuropeptide signaling pathway);GO:0010468(regulation of gene expression);GO:0048406(nerve growth factor binding);GO:0046323(glucose import);GO:0032868(response to insulin);GO:0008333(endosome to lysosome transport);GO:0010465(nerve growth factor receptor activity);GO:0030379(neurotensin receptor activity, non-G-protein coupled);GO:0006895(Golgi to endosome transport);GO:0014902(myotube differentiation);GO:0032509(endosome transport via multivesicular body sorting pathway);GO:0038180(nerve growth factor signaling pathway);GO:0045599(negative regulation of fat cell differentiation);GO:0048011(neurotrophin TRK receptor signaling pathway);GO:0048227(plasma membrane to endosome transport);GO:1904037(positive regulation of epithelial cell apoptotic process);GO:0030136(clathrin-coated vesicle);GO:0030140(trans-Golgi network transport vesicle);GO:0030425(dendrite);GO:0030659(cytoplasmic vesicle membrane);GO:0043025(neuronal cell body);GO:0043231(intracellular membrane-bounded organelle) | 04142(Lysosome);04979(Cholesterol metabolism);04722(Neurotrophin signaling pathway) | K12388 | NA | sortilin 1 [Source:HGNC Symbol;Acc:HGNC:11186] | 7.04 | 0.09 | 75.4645484376005 | 6.24 | 4.00e-14 | 3.65e-11 | up | yes |
| 78166 | NA | chr17 | - | 7665416 | 7687491 | ENST00000635293 | 12 | 1883 | ENSG00000141510 | TP53 | GO:0008285(negative regulation of cell proliferation);GO:0010628(positive regulation of gene expression);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0005524(ATP binding);GO:0005654(nucleoplasm);GO:0006355(regulation of transcription, DNA-templated);GO:0006366(transcription from RNA polymerase II promoter);GO:0006351(transcription, DNA-templated);GO:0003677(DNA binding);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0005829(cytosol);GO:0005783(endoplasmic reticulum);GO:0008270(zinc ion binding);GO:0005739(mitochondrion);GO:0006915(apoptotic process);GO:0005515(protein binding);GO:0006974(cellular response to DNA damage stimulus);GO:0007049(cell cycle);GO:0048511(rhythmic process);GO:0007275(multicellular organism development);GO:0030154(cell differentiation);GO:0005759(mitochondrial matrix);GO:0044212(transcription regulatory region DNA binding);GO:0051262(protein tetramerization);GO:0045893(positive regulation of transcription, DNA-templated);GO:0005730(nucleolus);GO:0042802(identical protein binding);GO:0006977(DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest);GO:0019901(protein kinase binding);GO:0016579(protein deubiquitination);GO:0043161(proteasome-mediated ubiquitin-dependent protein catabolic process);GO:0003682(chromatin binding);GO:0008283(cell proliferation);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0007050(cell cycle arrest);GO:0043065(positive regulation of apoptotic process);GO:0016032(viral process);GO:0031625(ubiquitin protein ligase binding);GO:0006284(base-excision repair);GO:0047485(protein N-terminus binding);GO:0050731(positive regulation of peptidyl-tyrosine phosphorylation);GO:0019899(enzyme binding);GO:0019903(protein phosphatase binding);GO:0008134(transcription factor binding);GO:0030308(negative regulation of cell growth);GO:0002020(protease binding);GO:0042981(regulation of apoptotic process);GO:0005507(copper ion binding);GO:0045892(negative regulation of transcription, DNA-templated);GO:0000790(nuclear chromatin);GO:0016363(nuclear matrix);GO:0046982(protein heterodimerization activity);GO:1901796(regulation of signal transduction by p53 class mediator);GO:0043234(protein complex);GO:0001085(RNA polymerase II transcription factor binding);GO:0016605(PML body);GO:0038111(interleukin-7-mediated signaling pathway);GO:0048015(phosphatidylinositol-mediated signaling);GO:0000981(RNA polymerase II transcription factor activity, sequence-specific DNA binding);GO:0035690(cellular response to drug);GO:0012501(programmed cell death);GO:0090200(positive regulation of release of cytochrome c from mitochondria);GO:2001244(positive regulation of intrinsic apoptotic signaling pathway);GO:0001046(core promoter sequence-specific DNA binding);GO:0051087(chaperone binding);GO:0090399(replicative senescence);GO:1902895(positive regulation of pri-miRNA transcription from RNA polymerase II promoter);GO:0006461(protein complex assembly);GO:0032461(positive regulation of protein oligomerization);GO:0007569(cell aging);GO:0071456(cellular response to hypoxia);GO:0002039(p53 binding);GO:0003684(damaged DNA binding);GO:0010165(response to X-ray);GO:0042771(intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator);GO:1900740(positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway);GO:0043153(entrainment of circadian clock by photoperiod);GO:0042149(cellular response to glucose starvation);GO:0071158(positive regulation of cell cycle arrest);GO:0072332(intrinsic apoptotic signaling pathway by p53 class mediator);GO:1990440(positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress);GO:0008104(protein localization);GO:0030330(DNA damage response, signal transduction by p53 class mediator);GO:0051721(protein phosphatase 2A binding);GO:0046677(response to antibiotic);GO:0000977(RNA polymerase II regulatory region sequence-specific DNA binding);GO:0097252(oligodendrocyte apoptotic process);GO:0048512(circadian behavior);GO:0003730(mRNA 3'-UTR binding);GO:0097718(disordered domain specific binding);GO:0035035(histone acetyltransferase binding);GO:0043066(negative regulation of apoptotic process);GO:2000379(positive regulation of reactive oxygen species metabolic process);GO:0048147(negative regulation of fibroblast proliferation);GO:0007265(Ras protein signal transduction);GO:0071480(cellular response to gamma radiation);GO:0005622(intracellular);GO:0005669(transcription factor TFIID complex);GO:0016604(nuclear body);GO:0000990(transcription factor activity, core RNA polymerase binding);GO:0001228(transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding);GO:0030971(receptor tyrosine kinase binding);GO:0042826(histone deacetylase binding);GO:0043621(protein self-association);GO:0000733(DNA strand renaturation);GO:0006289(nucleotide-excision repair);GO:0006914(autophagy);GO:0006978(DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator);GO:0006983(ER overload response);GO:0008340(determination of adult lifespan);GO:0010332(response to gamma radiation);GO:0031065(positive regulation of histone deacetylation);GO:0031497(chromatin assembly);GO:0031571(mitotic G1 DNA damage checkpoint);GO:0034644(cellular response to UV);GO:0043525(positive regulation of neuron apoptotic process);GO:0046827(positive regulation of protein export from nucleus);GO:0046902(regulation of mitochondrial membrane permeability);GO:0051097(negative regulation of helicase activity);GO:0051289(protein homotetramerization);GO:0051974(negative regulation of telomerase activity);GO:0070245(positive regulation of thymocyte apoptotic process);GO:0071479(cellular response to ionizing radiation);GO:0072331(signal transduction by p53 class mediator);GO:0072717(cellular response to actinomycin D);GO:0090403(oxidative stress-induced premature senescence);GO:0097193(intrinsic apoptotic signaling pathway);GO:1900119(positive regulation of execution phase of apoptosis);GO:1902749(regulation of cell cycle G2/M phase transition);GO:0005657(replication fork);GO:0016020(membrane);GO:0016021(integral component of membrane) | 04010(MAPK signaling pathway);04310(Wnt signaling pathway);04071(Sphingolipid signaling pathway);04151(PI3K-Akt signaling pathway);04137(Mitophagy - animal);04110(Cell cycle);04210(Apoptosis);04216(Ferroptosis);04115(p53 signaling pathway);04218(Cellular senescence);04919(Thyroid hormone signaling pathway);04722(Neurotrophin signaling pathway);04211(Longevity regulating pathway - mammal);05200(Pathways in cancer);05230(Central carbon metabolism in cancer);05202(Transcriptional misregulation in cancers);05206(MicroRNAs in cancer);05205(Proteoglycans in cancer);05203(Viral carcinogenesis);05210(Colorectal cancer);05212(Pancreatic cancer);05225(Hepatocellular carcinoma);05226(Gastric cancer);05214(Glioma);05216(Thyroid cancer);05220(Chronic myeloid leukemia);05217(Basal cell carcinoma);05218(Melanoma);05219(Bladder cancer);05215(Prostate cancer);05213(Endometrial cancer);05224(Breast cancer);05222(Small cell lung cancer);05223(Non-small cell lung cancer);05014(Amyotrophic lateral sclerosis (ALS));05016(Huntington's disease);05418(Fluid shear stress and atherosclerosis);05166(HTLV-I infection);05162(Measles);05161(Hepatitis B);05160(Hepatitis C);05168(Herpes simplex infection);05167(Kaposi's sarcoma-associated herpesvirus infection);05169(Epstein-Barr virus infection);05165(Human papillomavirus infection);01524(Platinum drug resistance);01522(Endocrine resistance) | NA | NA | tumor protein p53 [Source:HGNC Symbol;Acc:HGNC:11998] | 22.52117 | 0.31 | 73.1957787860272 | 6.19 | 6.51e-14 | 5.48e-11 | up | yes |
| 47619 | NA | chr12 | + | 123973123 | 124015422 | ENST00000539644 | 6 | 5108 | ENSG00000179195 | ZNF664 | GO:0005634(nucleus);GO:0003676(nucleic acid binding);GO:0046872(metal ion binding);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0003674(molecular_function);GO:0008150(biological_process) | NA | NA | NA | zinc finger protein 664 [Source:HGNC Symbol;Acc:HGNC:25406] | 0.47 | 33.705395 | 0.01 | -6.15657940729979 | 6.60e-14 | 5.48e-11 | down | yes |
| 43793 | NA | chr12 | + | 69585666 | 69601565 | ENST00000543146 | 16 | 2189 | ENSG00000166226 | CCT2 | GO:0005737(cytoplasm);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0005829(cytosol);GO:0006457(protein folding);GO:0005515(protein binding);GO:0005874(microtubule);GO:0051082(unfolded protein binding);GO:0005576(extracellular region);GO:0070062(extracellular exosome);GO:0031012(extracellular matrix);GO:0043312(neutrophil degranulation);GO:0035578(azurophil granule lumen);GO:0050821(protein stabilization);GO:0032212(positive regulation of telomere maintenance via telomerase);GO:0051973(positive regulation of telomerase activity);GO:0031625(ubiquitin protein ligase binding);GO:1904851(positive regulation of establishment of protein localization to telomere);GO:1904874(positive regulation of telomerase RNA localization to Cajal body);GO:0044183(protein binding involved in protein folding);GO:0090666(scaRNA localization to Cajal body);GO:1904871(positive regulation of protein localization to Cajal body);GO:0005832(chaperonin-containing T-complex);GO:0007339(binding of sperm to zona pellucida);GO:0051086(chaperone mediated protein folding independent of cofactor);GO:0051131(chaperone-mediated protein complex assembly);GO:1901998(toxin transport);GO:0002199(zona pellucida receptor complex);GO:0043209(myelin sheath);GO:0044297(cell body) | NA | NA | NA | chaperonin containing TCP1 subunit 2 [Source:HGNC Symbol;Acc:HGNC:1615] | 0.31 | 22.747316 | 0.01 | -6.21367485884939 | 7.50e-14 | 5.96e-11 | down | yes |
| 88861 | NA | chr17 | - | 82242667 | 82273731 | ENST00000314028 | 9 | 3712 | ENSG00000141551 | CSNK1D | GO:0000166(nucleotide binding);GO:0004672(protein kinase activity);GO:0004674(protein serine/threonine kinase activity);GO:0005524(ATP binding);GO:0006468(protein phosphorylation);GO:0001934(positive regulation of protein phosphorylation);GO:0090263(positive regulation of canonical Wnt signaling pathway);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0016740(transferase activity);GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0005794(Golgi apparatus);GO:0005886(plasma membrane);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0048511(rhythmic process);GO:0016055(Wnt signaling pathway);GO:0048471(perinuclear region of cytoplasm);GO:0005813(centrosome);GO:0005815(microtubule organizing center);GO:0005819(spindle);GO:0000139(Golgi membrane);GO:0005654(nucleoplasm);GO:0005829(cytosol);GO:0045296(cadherin binding);GO:0008360(regulation of cell shape);GO:0000086(G2/M transition of mitotic cell cycle);GO:0010389(regulation of G2/M transition of mitotic cell cycle);GO:0097711(ciliary basal body docking);GO:0005876(spindle microtubule);GO:0032922(circadian regulation of gene expression);GO:0006364(rRNA processing);GO:0048208(COPII vesicle coating);GO:0033116(endoplasmic reticulum-Golgi intermediate compartment membrane);GO:0042752(regulation of circadian rhythm);GO:0051225(spindle assembly);GO:0006897(endocytosis);GO:0032436(positive regulation of proteasomal ubiquitin-dependent protein catabolic process);GO:0018105(peptidyl-serine phosphorylation);GO:0050321(tau-protein kinase activity);GO:0042277(peptide binding);GO:0007020(microtubule nucleation);GO:0007030(Golgi organization);GO:0030177(positive regulation of Wnt signaling pathway);GO:0034067(protein localization to Golgi apparatus);GO:0061512(protein localization to cilium);GO:0071539(protein localization to centrosome);GO:1905426(positive regulation of Wnt-mediated midbrain dopaminergic neuron differentiation);GO:1905515(non-motile cilium assembly);GO:1990090(cellular response to nerve growth factor stimulus);GO:2000052(positive regulation of non-canonical Wnt signaling pathway);GO:0043005(neuron projection) | 04340(Hedgehog signaling pathway);04390(Hippo signaling pathway);04540(Gap junction);04710(Circadian rhythm) | K08959 | EC:2.7.11.1 | casein kinase 1 delta [Source:HGNC Symbol;Acc:HGNC:2452] | 0.40 | 27.161812 | 0.01 | -6.10163055546399 | 1.12e-13 | 8.54e-11 | down | yes |
| 105475 | NA | chr19 | + | 58386400 | 58394806 | ENST00000601521 | 7 | 1203 | ENSG00000083845 | RPS5 | GO:0003723(RNA binding);GO:0016020(membrane);GO:0006412(translation);GO:0006413(translational initiation);GO:0030529(intracellular ribonucleoprotein complex);GO:0003735(structural constituent of ribosome);GO:0005840(ribosome);GO:0015935(small ribosomal subunit);GO:0022627(cytosolic small ribosomal subunit);GO:0005925(focal adhesion);GO:0019843(rRNA binding);GO:0003729(mRNA binding);GO:0000028(ribosomal small subunit assembly);GO:0005654(nucleoplasm);GO:0005829(cytosol);GO:0070062(extracellular exosome);GO:0031012(extracellular matrix);GO:0005515(protein binding);GO:0006364(rRNA processing);GO:0000184(nuclear-transcribed mRNA catabolic process, nonsense-mediated decay);GO:0019083(viral transcription);GO:0006614(SRP-dependent cotranslational protein targeting to membrane);GO:0006450(regulation of translational fidelity) | 03010(Ribosome) | NA | NA | ribosomal protein S5 [Source:HGNC Symbol;Acc:HGNC:10426] | 42.727242 | 0.64 | 66.972803180042 | 6.07 | 1.25e-13 | 9.13e-11 | up | yes |
| 31117 | NA | chr11 | - | 65916808 | 65919117 | ENST00000438576 | 2 | 1619 | ENSG00000175573 | C11orf68 | GO:0003723(RNA binding);GO:0005515(protein binding) | NA | NA | NA | chromosome 11 open reading frame 68 [Source:HGNC Symbol;Acc:HGNC:28801] | 61.703274 | 0.96 | 64.0070144646103 | 6.00 | 1.30e-13 | 9.13e-11 | up | yes |
| 4956 | NA | chr1 | - | 43363398 | 43367738 | ENST00000621943 | 8 | 1625 | ENSG00000066322 | ELOVL1 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0016740(transferase activity);GO:0006629(lipid metabolic process);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0009922(fatty acid elongase activity);GO:0102336(3-oxo-arachidoyl-CoA synthase activity);GO:0102337(3-oxo-cerotoyl-CoA synthase activity);GO:0102338(3-oxo-lignoceronyl-CoA synthase activity);GO:0006631(fatty acid metabolic process);GO:0006633(fatty acid biosynthetic process);GO:0019367(fatty acid elongation, saturated fatty acid);GO:0042761(very long-chain fatty acid biosynthetic process);GO:0034625(fatty acid elongation, monounsaturated fatty acid);GO:0005515(protein binding);GO:0035338(long-chain fatty-acyl-CoA biosynthetic process);GO:0030176(integral component of endoplasmic reticulum membrane);GO:0036109(alpha-linolenic acid metabolic process);GO:0030148(sphingolipid biosynthetic process);GO:0043651(linoleic acid metabolic process);GO:0034626(fatty acid elongation, polyunsaturated fatty acid) | 00062(Fatty acid elongation) | K10247 | EC:2.3.1.199 | ELOVL fatty acid elongase 1 [Source:HGNC Symbol;Acc:HGNC:14418] | 1.28 | 84.109543 | 0.02 | -6.04175543223975 | 1.46e-13 | 9.84e-11 | down | yes |
| 77812 | NA | chr17 | - | 7235028 | 7239506 | ENST00000320316 | 5 | 2037 | ENSG00000040633 | PHF23 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0005654(nucleoplasm);GO:0006914(autophagy);GO:0003682(chromatin binding);GO:0005515(protein binding);GO:1902902(negative regulation of autophagosome assembly);GO:0007076(mitotic chromosome condensation);GO:0031398(positive regulation of protein ubiquitination);GO:1901097(negative regulation of autophagosome maturation) | NA | NA | NA | PHD finger protein 23 [Source:HGNC Symbol;Acc:HGNC:28428] | 23.344927 | 0.35 | 66.0982969265399 | 6.05 | 1.50e-13 | 9.84e-11 | up | yes |
| 191838 | NA | chr9 | + | 128203417 | 128255246 | ENST00000627543 | 22 | 4734 | ENSG00000106976 | DNM1 | GO:0003723(RNA binding);GO:0005737(cytoplasm);GO:0005525(GTP binding);GO:0000166(nucleotide binding);GO:0016787(hydrolase activity);GO:0003924(GTPase activity);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0045202(synapse);GO:0005874(microtubule);GO:0005886(plasma membrane);GO:0042802(identical protein binding);GO:0072583(clathrin-dependent endocytosis);GO:0006897(endocytosis);GO:0070062(extracellular exosome);GO:0008017(microtubule binding);GO:0048013(ephrin receptor signaling pathway);GO:0003374(dynamin family protein polymerization involved in mitochondrial fission);GO:0006898(receptor-mediated endocytosis);GO:0019901(protein kinase binding);GO:0061025(membrane fusion);GO:0031966(mitochondrial membrane);GO:0000266(mitochondrial fission);GO:0008022(protein C-terminus binding);GO:0031749(D2 dopamine receptor binding);GO:0032403(protein complex binding);GO:0046983(protein dimerization activity);GO:0050998(nitric-oxide synthase binding);GO:0002031(G-protein coupled receptor internalization);GO:0007032(endosome organization);GO:0007605(sensory perception of sound);GO:0008344(adult locomotory behavior);GO:0016185(synaptic vesicle budding from presynaptic endocytic zone membrane);GO:0031623(receptor internalization);GO:0051262(protein tetramerization);GO:0051932(synaptic transmission, GABAergic);GO:1901998(toxin transport);GO:1903423(positive regulation of synaptic vesicle recycling);GO:0001917(photoreceptor inner segment);GO:0005794(Golgi apparatus);GO:0008021(synaptic vesicle);GO:0030117(membrane coat);GO:0043196(varicosity);GO:0043209(myelin sheath);GO:0043234(protein complex) | 04072(Phospholipase D signaling pathway);04144(Endocytosis);04961(Endocrine and other factor-regulated calcium reabsorption);04721(Synaptic vesicle cycle);05100(Bacterial invasion of epithelial cells) | K01528 | EC:3.6.5.5 | dynamin 1 [Source:HGNC Symbol;Acc:HGNC:2972] | 0.12 | 8.11 | 0.01 | -6.11474594050073 | 1.86e-13 | 1.17e-10 | down | yes |
| 93192 | NA | chr19 | + | 1103926 | 1106787 | ENST00000616066 | 7 | 927 | ENSG00000167468 | GPX4 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0005654(nucleoplasm);GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0005739(mitochondrion);GO:0007275(multicellular organism development);GO:0098869(cellular oxidant detoxification);GO:0004601(peroxidase activity);GO:0004602(glutathione peroxidase activity);GO:0006979(response to oxidative stress);GO:0005829(cytosol);GO:0070062(extracellular exosome);GO:0043234(protein complex);GO:0006644(phospholipid metabolic process);GO:0019372(lipoxygenase pathway);GO:0042802(identical protein binding);GO:0047066(phospholipid-hydroperoxide glutathione peroxidase activity);GO:0051258(protein polymerization) | 00480(Glutathione metabolism);04216(Ferroptosis) | NA | NA | glutathione peroxidase 4 [Source:HGNC Symbol;Acc:HGNC:4556] | 71.248787 | 1.17 | 60.9146214679605 | 5.93 | 2.47e-13 | 1.50e-10 | up | yes |
| 47492 | NA | chr12 | - | 123260971 | 123268254 | ENST00000618072 | 4 | 1144 | ENSG00000111328 | CDK2AP1 | GO:0005634(nucleus);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0005515(protein binding);GO:0007049(cell cycle);GO:0048471(perinuclear region of cytoplasm);GO:0001934(positive regulation of protein phosphorylation);GO:0070182(DNA polymerase binding);GO:0006261(DNA-dependent DNA replication) | NA | NA | NA | cyclin dependent kinase 2 associated protein 1 [Source:HGNC Symbol;Acc:HGNC:14002] | 74.121292 | 1.30 | 56.8822675768285 | 5.83 | 4.24e-13 | 2.50e-10 | up | yes |
| 198694 | NA | chrX | + | 151397204 | 151409364 | ENST00000330374 | 3 | 4717 | ENSG00000160131 | VMA21 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0031410(cytoplasmic vesicle);GO:0070072(vacuolar proton-transporting V-type ATPase complex assembly);GO:0033116(endoplasmic reticulum-Golgi intermediate compartment membrane);GO:0012507(ER to Golgi transport vesicle membrane);GO:0043462(regulation of ATPase activity);GO:0005764(lysosome) | NA | NA | NA | VMA21, vacuolar ATPase assembly factor [Source:HGNC Symbol;Acc:HGNC:22082] | 0.12 | 7.53 | 0.02 | -5.94087101459259 | 5.27e-13 | 2.94e-10 | down | yes |
| 30214 | NA | chr11 | + | 63838928 | 63911019 | ENST00000402010 | 19 | 4728 | ENSG00000072518 | MARK2 | GO:0003723(RNA binding);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0046872(metal ion binding);GO:0000166(nucleotide binding);GO:0004672(protein kinase activity);GO:0004674(protein serine/threonine kinase activity);GO:0005524(ATP binding);GO:0006468(protein phosphorylation);GO:0005654(nucleoplasm);GO:0016740(transferase activity);GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0005886(plasma membrane);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0007275(multicellular organism development);GO:0016055(Wnt signaling pathway);GO:0000287(magnesium ion binding);GO:0030154(cell differentiation);GO:0000226(microtubule cytoskeleton organization);GO:0030010(establishment of cell polarity);GO:0008289(lipid binding);GO:0045296(cadherin binding);GO:0016328(lateral plasma membrane);GO:0046777(protein autophosphorylation);GO:0005739(mitochondrion);GO:0035556(intracellular signal transduction);GO:0005884(actin filament);GO:0051493(regulation of cytoskeleton organization);GO:0050770(regulation of axonogenesis);GO:0010976(positive regulation of neuron projection development);GO:0001764(neuron migration);GO:0097427(microtubule bundle);GO:0000422(mitophagy);GO:0018107(peptidyl-threonine phosphorylation);GO:0050321(tau-protein kinase activity);GO:0045197(establishment or maintenance of epithelial cell apical/basal polarity);GO:0032147(activation of protein kinase activity);GO:0051646(mitochondrion localization);GO:0030295(protein kinase activator activity) | NA | NA | NA | microtubule affinity regulating kinase 2 [Source:HGNC Symbol;Acc:HGNC:3332] | 10.549787 | 0.18 | 57.2334629928009 | 5.84 | 5.32e-13 | 2.94e-10 | up | yes |
| 14594 | NA | chr1 | - | 201483532 | 201492499 | ENST00000533402 | 2 | 8475 | ENSG00000159176 | CSRP1 | GO:0003723(RNA binding);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0005925(focal adhesion);GO:0070062(extracellular exosome);GO:0070527(platelet aggregation);GO:0008270(zinc ion binding) | NA | NA | NA | cysteine and glycine rich protein 1 [Source:HGNC Symbol;Acc:HGNC:2469] | 22.956362 | 0.45 | 51.0670205145773 | 5.67 | 9.71e-13 | 5.19e-10 | up | yes |
| 154605 | NA | chr5 | - | 78485217 | 78648825 | ENST00000380345 | 5 | 5043 | ENSG00000145685 | LHFPL2 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005886(plasma membrane);GO:0003674(molecular_function);GO:0008150(biological_process);GO:0005575(cellular_component);GO:0002576(platelet degranulation);GO:0031092(platelet alpha granule membrane) | NA | NA | NA | LHFPL tetraspan subfamily member 2 [Source:HGNC Symbol;Acc:HGNC:6588] | 7.01 | 0.13 | 53.8550001537563 | 5.75 | 9.95e-13 | 5.19e-10 | up | yes |
| 164710 | NA | chr6 | + | 44246166 | 44253888 | ENST00000620073 | 12 | 2644 | ENSG00000096384 | HSP90AB1 | GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0005829(cytosol);GO:0006457(protein folding);GO:0005739(mitochondrion);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0005576(extracellular region);GO:0042470(melanosome);GO:0051082(unfolded protein binding);GO:0006950(response to stress);GO:0038096(Fc-gamma receptor signaling pathway involved in phagocytosis);GO:0006986(response to unfolded protein);GO:0005654(nucleoplasm);GO:0045296(cadherin binding);GO:0070062(extracellular exosome);GO:0008180(COP9 signalosome);GO:0071157(negative regulation of cell cycle arrest);GO:0043234(protein complex);GO:0006805(xenobiotic metabolic process);GO:0043312(neutrophil degranulation);GO:0032435(negative regulation of proteasomal ubiquitin-dependent protein catabolic process);GO:0034774(secretory granule lumen);GO:1904813(ficolin-1-rich granule lumen);GO:0045597(positive regulation of cell differentiation);GO:0030511(positive regulation of transforming growth factor beta receptor signaling pathway);GO:0030911(TPR domain binding);GO:1990226(histone methyltransferase binding);GO:0051973(positive regulation of telomerase activity);GO:0007004(telomere maintenance via telomerase);GO:0042277(peptide binding);GO:0042826(histone deacetylase binding);GO:1900034(regulation of cellular response to heat);GO:0019900(kinase binding);GO:0003725(double-stranded RNA binding);GO:0042803(protein homodimerization activity);GO:0046983(protein dimerization activity);GO:0031072(heat shock protein binding);GO:0097718(disordered domain specific binding);GO:0030235(nitric-oxide synthase regulator activity);GO:0045429(positive regulation of nitric oxide biosynthetic process);GO:0023026(MHC class II protein complex binding);GO:0070182(DNA polymerase binding);GO:0031396(regulation of protein ubiquitination);GO:0050821(protein stabilization);GO:1905323(telomerase holoenzyme complex assembly);GO:0043008(ATP-dependent protein binding);GO:0019062(virion attachment to host cell);GO:0032516(positive regulation of phosphoprotein phosphatase activity);GO:0051131(chaperone-mediated protein complex assembly);GO:0060334(regulation of interferon-gamma-mediated signaling pathway);GO:0060338(regulation of type I interferon-mediated signaling pathway);GO:0097435(supramolecular fiber organization);GO:1901389(negative regulation of transforming growth factor beta activation);GO:2000010(positive regulation of protein localization to cell surface);GO:0034751(aryl hydrocarbon receptor complex);GO:0002134(UTP binding);GO:0002135(CTP binding);GO:0005525(GTP binding);GO:0008144(drug binding);GO:0017098(sulfonylurea receptor binding);GO:0019887(protein kinase regulator activity);GO:0019901(protein kinase binding);GO:0032564(dATP binding);GO:0044325(ion channel binding);GO:0001890(placenta development);GO:0009651(response to salt stress);GO:0010033(response to organic substance);GO:0032092(positive regulation of protein binding);GO:0033160(positive regulation of protein import into nucleus, translocation);GO:0035690(cellular response to drug);GO:0042220(response to cocaine);GO:0042493(response to drug);GO:0043066(negative regulation of apoptotic process);GO:0043524(negative regulation of neuron apoptotic process);GO:0045793(positive regulation of cell size);GO:0071353(cellular response to interleukin-4);GO:0071407(cellular response to organic cyclic compound);GO:0071902(positive regulation of protein serine/threonine kinase activity);GO:1903660(negative regulation of complement-dependent cytotoxicity);GO:0005622(intracellular);GO:0005765(lysosomal membrane);GO:0009986(cell surface);GO:0016234(inclusion body);GO:0016323(basolateral plasma membrane);GO:0016324(apical plasma membrane);GO:0031526(brush border membrane);GO:1990913(sperm head plasma membrane);GO:1990917(ooplasm) | 04141(Protein processing in endoplasmic reticulum);04151(PI3K-Akt signaling pathway);04217(Necroptosis);04621(NOD-like receptor signaling pathway);04612(Antigen processing and presentation);04659(Th17 cell differentiation);04657(IL-17 signaling pathway);04915(Estrogen signaling pathway);04914(Progesterone-mediated oocyte maturation);05200(Pathways in cancer);05215(Prostate cancer);05418(Fluid shear stress and atherosclerosis) | K04079 | NA | heat shock protein 90 alpha family class B member 1 [Source:HGNC Symbol;Acc:HGNC:5258] | 2.53 | 132.621124 | 0.02 | -5.71474626836481 | 1.12e-12 | 5.68e-10 | down | yes |
| 95387 | NA | chr19 | - | 10316212 | 10333546 | ENST00000617231 | 13 | 3501 | ENSG00000161847 | RAVER1 | GO:0003676(nucleic acid binding);GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0000398(mRNA splicing, via spliceosome) | NA | NA | NA | ribonucleoprotein, PTB binding 1 [Source:HGNC Symbol;Acc:HGNC:30296] | 10.025638 | 0.19 | 53.3664672316144 | 5.74 | 1.20e-12 | 5.95e-10 | up | yes |
| 116243 | NA | chr2 | + | 191245389 | 191425384 | ENST00000392318 | 31 | 5082 | ENSG00000128641 | MYO1B | GO:0005524(ATP binding);GO:0003774(motor activity);GO:0016459(myosin complex);GO:0005737(cytoplasm);GO:0000166(nucleotide binding);GO:0003779(actin binding);GO:0005886(plasma membrane);GO:0048471(perinuclear region of cytoplasm);GO:0005516(calmodulin binding);GO:0045296(cadherin binding);GO:0051015(actin filament binding);GO:0030175(filopodium);GO:0070062(extracellular exosome);GO:0071944(cell periphery);GO:0005769(early endosome);GO:0010008(endosome membrane);GO:0005546(phosphatidylinositol-4,5-bisphosphate binding);GO:0005547(phosphatidylinositol-3,4,5-trisphosphate binding);GO:0005903(brush border);GO:0051017(actin filament bundle assembly);GO:0005884(actin filament);GO:0007015(actin filament organization);GO:0032588(trans-Golgi network membrane);GO:0000146(microfilament motor activity);GO:0006892(post-Golgi vesicle-mediated transport);GO:0030048(actin filament-based movement);GO:0030898(actin-dependent ATPase activity);GO:0045177(apical part of cell) | NA | NA | NA | myosin IB [Source:HGNC Symbol;Acc:HGNC:7596] | 11.281031 | 0.22 | 50.3473605755498 | 5.65 | 1.67e-12 | 8.06e-10 | up | yes |
| 173230 | NA | chr7 | + | 73830863 | 73832693 | ENST00000340958 | 1 | 1831 | ENSG00000189143 | CLDN4 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005198(structural molecule activity);GO:0005886(plasma membrane);GO:0030054(cell junction);GO:0007165(signal transduction);GO:0005923(bicellular tight junction);GO:0005887(integral component of plasma membrane);GO:0042802(identical protein binding);GO:0016338(calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules);GO:0004888(transmembrane signaling receptor activity);GO:0007565(female pregnancy);GO:0007623(circadian rhythm);GO:0032570(response to progesterone);GO:0061436(establishment of skin barrier);GO:0009925(basal plasma membrane);GO:0016324(apical plasma membrane);GO:0016327(apicolateral plasma membrane);GO:0016328(lateral plasma membrane) | 04514(Cell adhesion molecules (CAMs));04530(Tight junction);04670(Leukocyte transendothelial migration);05160(Hepatitis C) | K06087 | NA | claudin 4 [Source:HGNC Symbol;Acc:HGNC:2046] | 2.13 | 105.456154 | 0.02 | -5.63159466185974 | 2.04e-12 | 9.55e-10 | down | yes |
| 132799 | NA | chr3 | - | 25597905 | 25664372 | ENST00000264331 | 36 | 5358 | ENSG00000077097 | TOP2B | GO:0005524(ATP binding);GO:0003677(DNA binding);GO:0006265(DNA topological change);GO:0003918(DNA topoisomerase type II (ATP-hydrolyzing) activity);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0000166(nucleotide binding);GO:0016853(isomerase activity);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0006259(DNA metabolic process);GO:0003916(DNA topoisomerase activity);GO:0000712(resolution of meiotic recombination intermediates);GO:0003682(chromatin binding);GO:0008022(protein C-terminus binding);GO:0019899(enzyme binding);GO:0046982(protein heterodimerization activity);GO:0042826(histone deacetylase binding);GO:0016925(protein sumoylation);GO:0000792(heterochromatin);GO:0005080(protein kinase C binding);GO:0000819(sister chromatid segregation);GO:0044774(mitotic DNA integrity checkpoint);GO:0001764(neuron migration);GO:0007409(axonogenesis);GO:0030900(forebrain development) | 01524(Platinum drug resistance) | K03164 | EC:5.99.1.3 | topoisomerase (DNA) II beta [Source:HGNC Symbol;Acc:HGNC:11990] | 5.62 | 0.11 | 50.0923571702198 | 5.65 | 2.35e-12 | 1.07e-09 | up | yes |
| 110591 | NA | chr2 | + | 85539642 | 85543608 | ENST00000409017 | 8 | 1927 | ENSG00000168906 | MAT2A | GO:0046872(metal ion binding);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0016740(transferase activity);GO:0006730(one-carbon metabolic process);GO:0005515(protein binding);GO:0042802(identical protein binding);GO:0004478(methionine adenosyltransferase activity);GO:0006556(S-adenosylmethionine biosynthetic process);GO:1990830(cellular response to leukemia inhibitory factor);GO:0005829(cytosol);GO:0051291(protein heterooligomerization);GO:0034214(protein hexamerization);GO:0032259(methylation);GO:0048269(methionine adenosyltransferase complex) | 00270(Cysteine and methionine metabolism) | NA | NA | methionine adenosyltransferase 2A [Source:HGNC Symbol;Acc:HGNC:6904] | 0.68 | 33.320889 | 0.02 | -5.60460880728083 | 3.08e-12 | 1.37e-09 | down | yes |
| 93198 | NA | chr19 | + | 1104415 | 1105761 | ENST00000585362 | 4 | 859 | ENSG00000167468 | GPX4 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0005654(nucleoplasm);GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0005739(mitochondrion);GO:0007275(multicellular organism development);GO:0098869(cellular oxidant detoxification);GO:0004601(peroxidase activity);GO:0004602(glutathione peroxidase activity);GO:0006979(response to oxidative stress);GO:0005829(cytosol);GO:0070062(extracellular exosome);GO:0043234(protein complex);GO:0006644(phospholipid metabolic process);GO:0019372(lipoxygenase pathway);GO:0042802(identical protein binding);GO:0047066(phospholipid-hydroperoxide glutathione peroxidase activity);GO:0051258(protein polymerization) | 00480(Glutathione metabolism);04216(Ferroptosis) | NA | NA | glutathione peroxidase 4 [Source:HGNC Symbol;Acc:HGNC:4556] | 65.268692 | 1.41 | 46.3012471863643 | 5.53 | 3.48e-12 | 1.51e-09 | up | yes |
| 122315 | NA | chr20 | + | 37296896 | 37317260 | ENST00000373605 | 4 | 1915 | ENSG00000101363 | MANBAL | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005515(protein binding) | NA | NA | NA | mannosidase beta like [Source:HGNC Symbol;Acc:HGNC:15799] | 0.74 | 35.409195 | 0.02 | -5.57116590343919 | 3.85e-12 | 1.63e-09 | down | yes |
| 22868 | NA | chr10 | - | 102479229 | 102502711 | ENST00000369905 | 11 | 2829 | ENSG00000138107 | ACTR1A | GO:0005869(dynactin complex);GO:0005737(cytoplasm);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0015630(microtubule cytoskeleton);GO:0005856(cytoskeleton);GO:0005813(centrosome);GO:0005815(microtubule organizing center);GO:0005515(protein binding);GO:0005829(cytosol);GO:0070062(extracellular exosome);GO:0005938(cell cortex);GO:0000086(G2/M transition of mitotic cell cycle);GO:0010389(regulation of G2/M transition of mitotic cell cycle);GO:0097711(ciliary basal body docking);GO:0019886(antigen processing and presentation of exogenous peptide antigen via MHC class II);GO:0016192(vesicle-mediated transport);GO:0006888(ER to Golgi vesicle-mediated transport);GO:0005875(microtubule associated complex);GO:0099738(cell cortex region);GO:0007283(spermatogenesis);GO:0002177(manchette);GO:0005814(centriole);GO:0030137(COPI-coated vesicle);GO:0043209(myelin sheath) | NA | NA | NA | ARP1 actin related protein 1 homolog A [Source:HGNC Symbol;Acc:HGNC:167] | 81.470779 | 1.91 | 42.6328051248883 | 5.41 | 5.41e-12 | 2.24e-09 | up | yes |
| 106313 | NA | chr2 | + | 11677595 | 11824887 | ENST00000449576 | 22 | 3077 | ENSG00000134324 | LPIN1 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0016787(hydrolase activity);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0006357(regulation of transcription from RNA polymerase II promoter);GO:0006629(lipid metabolic process);GO:0005829(cytosol);GO:0008195(phosphatidate phosphatase activity);GO:0016311(dephosphorylation);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0006631(fatty acid metabolic process);GO:0019432(triglyceride biosynthetic process);GO:0009062(fatty acid catabolic process);GO:0003674(molecular_function);GO:0005654(nucleoplasm);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0031965(nuclear membrane);GO:0007077(mitotic nuclear envelope disassembly);GO:0003713(transcription coactivator activity);GO:0005635(nuclear envelope);GO:0006656(phosphatidylcholine biosynthetic process);GO:0006646(phosphatidylethanolamine biosynthetic process);GO:0005741(mitochondrial outer membrane);GO:0006642(triglyceride mobilization);GO:0031100(animal organ regeneration);GO:0032869(cellular response to insulin stimulus) | 00561(Glycerolipid metabolism);00564(Glycerophospholipid metabolism);04150(mTOR signaling pathway) | K15728 | EC:3.1.3.4 | lipin 1 [Source:HGNC Symbol;Acc:HGNC:13345] | 8.75 | 0.20 | 44.7262889813953 | 5.48 | 6.96e-12 | 2.72e-09 | up | yes |
| 125017 | NA | chr21 | - | 25880550 | 26170654 | ENST00000346798 | 18 | 3467 | ENSG00000142192 | APP | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0046872(metal ion binding);GO:0003677(DNA binding);GO:0007155(cell adhesion);GO:0005794(Golgi apparatus);GO:0004867(serine-type endopeptidase inhibitor activity);GO:0010951(negative regulation of endopeptidase activity);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0007399(nervous system development);GO:0006915(apoptotic process);GO:0048471(perinuclear region of cytoplasm);GO:0005615(extracellular space);GO:0007219(Notch signaling pathway);GO:0005790(smooth endoplasmic reticulum);GO:0005905(clathrin-coated pit);GO:0008201(heparin binding);GO:0005576(extracellular region);GO:0030414(peptidase inhibitor activity);GO:0010466(negative regulation of peptidase activity);GO:0042802(identical protein binding);GO:0046914(transition metal ion binding);GO:0016504(peptidase activator activity);GO:0070851(growth factor receptor binding);GO:0001967(suckling behavior);GO:0006378(mRNA polyadenylation);GO:0006417(regulation of translation);GO:0006468(protein phosphorylation);GO:0006878(cellular copper ion homeostasis);GO:0006897(endocytosis);GO:0006979(response to oxidative stress);GO:0007176(regulation of epidermal growth factor-activated receptor activity);GO:0007409(axonogenesis);GO:0007617(mating behavior);GO:0007626(locomotory behavior);GO:0008088(axo-dendritic transport);GO:0008203(cholesterol metabolic process);GO:0008344(adult locomotory behavior);GO:0008542(visual learning);GO:0010288(response to lead ion);GO:0010468(regulation of gene expression);GO:0010952(positive regulation of peptidase activity);GO:0010971(positive regulation of G2/M transition of mitotic cell cycle);GO:0016199(axon midline choice point recognition);GO:0016322(neuron remodeling);GO:0016358(dendrite development);GO:0030198(extracellular matrix organization);GO:0030900(forebrain development);GO:0031175(neuron projection development);GO:0035235(ionotropic glutamate receptor signaling pathway);GO:0040014(regulation of multicellular organism growth);GO:0043393(regulation of protein binding);GO:0045665(negative regulation of neuron differentiation);GO:0045931(positive regulation of mitotic cell cycle);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0048669(collateral sprouting in absence of injury);GO:0050803(regulation of synapse structure or activity);GO:0050885(neuromuscular process controlling balance);GO:0051124(synaptic growth at neuromuscular junction);GO:0051247(positive regulation of protein metabolic process);GO:0051402(neuron apoptotic process);GO:0051563(smooth endoplasmic reticulum calcium ion homeostasis);GO:0071320(cellular response to cAMP);GO:0071874(cellular response to norepinephrine stimulus);GO:1990090(cellular response to nerve growth factor stimulus);GO:0005791(rough endoplasmic reticulum);GO:0005911(cell-cell junction);GO:0009986(cell surface);GO:0030134(ER to Golgi transport vesicle);GO:0030424(axon);GO:0030426(growth cone);GO:0031410(cytoplasmic vesicle);GO:0031594(neuromuscular junction);GO:0035253(ciliary rootlet);GO:0043005(neuron projection);GO:0043195(terminal bouton);GO:0044304(main axon);GO:0045177(apical part of cell);GO:0051233(spindle midzone);GO:0097449(astrocyte projection);GO:1990761(growth cone lamellipodium);GO:1990812(growth cone filopodium);GO:0005768(endosome);GO:0005829(cytosol);GO:0070062(extracellular exosome);GO:0044267(cellular protein metabolic process);GO:0008285(negative regulation of cell proliferation);GO:0043687(post-translational protein modification);GO:0045087(innate immune response);GO:0005887(integral component of plasma membrane);GO:0031904(endosome lumen);GO:0032588(trans-Golgi network membrane);GO:0002576(platelet degranulation);GO:0010628(positive regulation of gene expression);GO:1904646(cellular response to amyloid-beta);GO:0005788(endoplasmic reticulum lumen);GO:0031093(platelet alpha granule lumen);GO:0005796(Golgi lumen);GO:0005102(receptor binding);GO:0043235(receptor complex);GO:0019899(enzyme binding);GO:0014005(microglia development);GO:0032640(tumor necrosis factor production);GO:0043197(dendritic spine);GO:0045202(synapse);GO:0007611(learning or memory);GO:0090647(modulation of age-related behavioral decline);GO:1900273(positive regulation of long-term synaptic potentiation);GO:0046330(positive regulation of JNK cascade);GO:0051425(PTB domain binding);GO:0001934(positive regulation of protein phosphorylation);GO:0002265(astrocyte activation involved in immune response);GO:0009987(cellular process);GO:0010629(negative regulation of gene expression);GO:0048143(astrocyte activation);GO:0048169(regulation of long-term neuronal synaptic plasticity);GO:0050808(synapse organization);GO:0051091(positive regulation of sequence-specific DNA binding transcription factor activity);GO:0098815(modulation of excitatory postsynaptic potential);GO:1990000(amyloid fibril formation);GO:2000310(regulation of NMDA receptor activity);GO:0005641(nuclear envelope lumen);GO:0043198(dendritic shaft);GO:0045121(membrane raft) | 04726(Serotonergic synapse);05010(Alzheimer's disease) | K04520 | NA | amyloid beta precursor protein [Source:HGNC Symbol;Acc:HGNC:620] | 0.38 | 16.896006 | 0.02 | -5.48387836710971 | 6.98e-12 | 2.72e-09 | down | yes |
| 167305 | NA | chr6 | - | 127288710 | 127343609 | ENST00000454859 | 6 | 2341 | ENSG00000093144 | ECHDC1 | GO:0005737(cytoplasm);GO:0003824(catalytic activity);GO:0016829(lyase activity);GO:0008152(metabolic process);GO:0005829(cytosol);GO:0016831(carboxy-lyase activity);GO:0005739(mitochondrion);GO:0070062(extracellular exosome);GO:0006635(fatty acid beta-oxidation);GO:0004300(enoyl-CoA hydratase activity);GO:0004492(methylmalonyl-CoA decarboxylase activity);GO:0016020(membrane);GO:0016021(integral component of membrane) | 00640(Propanoate metabolism) | K18426 | EC:4.1.1.94 4.1.1.41 | ethylmalonyl-CoA decarboxylase 1 [Source:HGNC Symbol;Acc:HGNC:21489] | 13.804155 | 0.31 | 44.489204946484 | 5.48 | 7.02e-12 | 2.72e-09 | up | yes |
| 118082 | NA | chr2 | + | 219178217 | 219185479 | ENST00000430297 | 9 | 4618 | ENSG00000144567 | RETREG2 | GO:0016020(membrane);GO:0016021(integral component of membrane) | NA | NA | NA | reticulophagy regulator family member 2 [Source:HGNC Symbol;Acc:HGNC:28450] | 15.906613 | 0.38 | 42.1180908048625 | 5.40 | 7.37e-12 | 2.80e-09 | up | yes |
| 78177 | NA | chr17 | - | 7668402 | 7687550 | ENST00000617185 | 12 | 2724 | ENSG00000141510 | TP53 | GO:0008285(negative regulation of cell proliferation);GO:0010628(positive regulation of gene expression);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0005524(ATP binding);GO:0005654(nucleoplasm);GO:0006355(regulation of transcription, DNA-templated);GO:0006366(transcription from RNA polymerase II promoter);GO:0006351(transcription, DNA-templated);GO:0003677(DNA binding);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0005829(cytosol);GO:0005783(endoplasmic reticulum);GO:0008270(zinc ion binding);GO:0005739(mitochondrion);GO:0006915(apoptotic process);GO:0005515(protein binding);GO:0006974(cellular response to DNA damage stimulus);GO:0007049(cell cycle);GO:0048511(rhythmic process);GO:0007275(multicellular organism development);GO:0030154(cell differentiation);GO:0005759(mitochondrial matrix);GO:0044212(transcription regulatory region DNA binding);GO:0051262(protein tetramerization);GO:0045893(positive regulation of transcription, DNA-templated);GO:0005730(nucleolus);GO:0042802(identical protein binding);GO:0006977(DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest);GO:0019901(protein kinase binding);GO:0016579(protein deubiquitination);GO:0043161(proteasome-mediated ubiquitin-dependent protein catabolic process);GO:0003682(chromatin binding);GO:0008283(cell proliferation);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0007050(cell cycle arrest);GO:0043065(positive regulation of apoptotic process);GO:0016032(viral process);GO:0031625(ubiquitin protein ligase binding);GO:0006284(base-excision repair);GO:0047485(protein N-terminus binding);GO:0050731(positive regulation of peptidyl-tyrosine phosphorylation);GO:0019899(enzyme binding);GO:0019903(protein phosphatase binding);GO:0008134(transcription factor binding);GO:0030308(negative regulation of cell growth);GO:0002020(protease binding);GO:0042981(regulation of apoptotic process);GO:0005507(copper ion binding);GO:0045892(negative regulation of transcription, DNA-templated);GO:0000790(nuclear chromatin);GO:0016363(nuclear matrix);GO:0046982(protein heterodimerization activity);GO:1901796(regulation of signal transduction by p53 class mediator);GO:0043234(protein complex);GO:0001085(RNA polymerase II transcription factor binding);GO:0016605(PML body);GO:0038111(interleukin-7-mediated signaling pathway);GO:0048015(phosphatidylinositol-mediated signaling);GO:0000981(RNA polymerase II transcription factor activity, sequence-specific DNA binding);GO:0035690(cellular response to drug);GO:0012501(programmed cell death);GO:0090200(positive regulation of release of cytochrome c from mitochondria);GO:2001244(positive regulation of intrinsic apoptotic signaling pathway);GO:0001046(core promoter sequence-specific DNA binding);GO:0051087(chaperone binding);GO:0090399(replicative senescence);GO:1902895(positive regulation of pri-miRNA transcription from RNA polymerase II promoter);GO:0006461(protein complex assembly);GO:0032461(positive regulation of protein oligomerization);GO:0007569(cell aging);GO:0071456(cellular response to hypoxia);GO:0002039(p53 binding);GO:0003684(damaged DNA binding);GO:0010165(response to X-ray);GO:0042771(intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator);GO:1900740(positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway);GO:0043153(entrainment of circadian clock by photoperiod);GO:0042149(cellular response to glucose starvation);GO:0071158(positive regulation of cell cycle arrest);GO:0072332(intrinsic apoptotic signaling pathway by p53 class mediator);GO:1990440(positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress);GO:0008104(protein localization);GO:0030330(DNA damage response, signal transduction by p53 class mediator);GO:0051721(protein phosphatase 2A binding);GO:0046677(response to antibiotic);GO:0000977(RNA polymerase II regulatory region sequence-specific DNA binding);GO:0097252(oligodendrocyte apoptotic process);GO:0048512(circadian behavior);GO:0003730(mRNA 3'-UTR binding);GO:0097718(disordered domain specific binding);GO:0035035(histone acetyltransferase binding);GO:0043066(negative regulation of apoptotic process);GO:2000379(positive regulation of reactive oxygen species metabolic process);GO:0048147(negative regulation of fibroblast proliferation);GO:0007265(Ras protein signal transduction);GO:0071480(cellular response to gamma radiation);GO:0005622(intracellular);GO:0005669(transcription factor TFIID complex);GO:0016604(nuclear body);GO:0000990(transcription factor activity, core RNA polymerase binding);GO:0001228(transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding);GO:0030971(receptor tyrosine kinase binding);GO:0042826(histone deacetylase binding);GO:0043621(protein self-association);GO:0000733(DNA strand renaturation);GO:0006289(nucleotide-excision repair);GO:0006914(autophagy);GO:0006978(DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator);GO:0006983(ER overload response);GO:0008340(determination of adult lifespan);GO:0010332(response to gamma radiation);GO:0031065(positive regulation of histone deacetylation);GO:0031497(chromatin assembly);GO:0031571(mitotic G1 DNA damage checkpoint);GO:0034644(cellular response to UV);GO:0043525(positive regulation of neuron apoptotic process);GO:0046827(positive regulation of protein export from nucleus);GO:0046902(regulation of mitochondrial membrane permeability);GO:0051097(negative regulation of helicase activity);GO:0051289(protein homotetramerization);GO:0051974(negative regulation of telomerase activity);GO:0070245(positive regulation of thymocyte apoptotic process);GO:0071479(cellular response to ionizing radiation);GO:0072331(signal transduction by p53 class mediator);GO:0072717(cellular response to actinomycin D);GO:0090403(oxidative stress-induced premature senescence);GO:0097193(intrinsic apoptotic signaling pathway);GO:1900119(positive regulation of execution phase of apoptosis);GO:1902749(regulation of cell cycle G2/M phase transition);GO:0005657(replication fork);GO:0016020(membrane);GO:0016021(integral component of membrane) | 04010(MAPK signaling pathway);04310(Wnt signaling pathway);04071(Sphingolipid signaling pathway);04151(PI3K-Akt signaling pathway);04137(Mitophagy - animal);04110(Cell cycle);04210(Apoptosis);04216(Ferroptosis);04115(p53 signaling pathway);04218(Cellular senescence);04919(Thyroid hormone signaling pathway);04722(Neurotrophin signaling pathway);04211(Longevity regulating pathway - mammal);05200(Pathways in cancer);05230(Central carbon metabolism in cancer);05202(Transcriptional misregulation in cancers);05206(MicroRNAs in cancer);05205(Proteoglycans in cancer);05203(Viral carcinogenesis);05210(Colorectal cancer);05212(Pancreatic cancer);05225(Hepatocellular carcinoma);05226(Gastric cancer);05214(Glioma);05216(Thyroid cancer);05220(Chronic myeloid leukemia);05217(Basal cell carcinoma);05218(Melanoma);05219(Bladder cancer);05215(Prostate cancer);05213(Endometrial cancer);05224(Breast cancer);05222(Small cell lung cancer);05223(Non-small cell lung cancer);05014(Amyotrophic lateral sclerosis (ALS));05016(Huntington's disease);05418(Fluid shear stress and atherosclerosis);05166(HTLV-I infection);05162(Measles);05161(Hepatitis B);05160(Hepatitis C);05168(Herpes simplex infection);05167(Kaposi's sarcoma-associated herpesvirus infection);05169(Epstein-Barr virus infection);05165(Human papillomavirus infection);01524(Platinum drug resistance);01522(Endocrine resistance) | K04451 | NA | tumor protein p53 [Source:HGNC Symbol;Acc:HGNC:11998] | 1.08 | 46.874588 | 0.02 | -5.43880806485692 | 7.66e-12 | 2.85e-09 | down | yes |
| 83274 | NA | chr17 | - | 44319625 | 44324870 | ENST00000225308 | 12 | 1607 | ENSG00000013306 | SLC25A39 | GO:0006810(transport);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005743(mitochondrial inner membrane);GO:0005739(mitochondrion);GO:0055085(transmembrane transport);GO:0006783(heme biosynthetic process);GO:0006839(mitochondrial transport);GO:0022857(transmembrane transporter activity) | NA | NA | NA | solute carrier family 25 member 39 [Source:HGNC Symbol;Acc:HGNC:24279] | 1.02 | 44.631493 | 0.02 | -5.45312921072762 | 8.13e-12 | 2.96e-09 | down | yes |
| 28445 | NA | chr11 | + | 47269376 | 47330031 | ENST00000342922 | 33 | 6005 | ENSG00000110514 | MADD | GO:0005829(cytosol);GO:0005886(plasma membrane);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0043547(positive regulation of GTPase activity);GO:0005515(protein binding);GO:0006915(apoptotic process);GO:0005085(guanyl-nucleotide exchange factor activity);GO:0017112(Rab guanyl-nucleotide exchange factor activity);GO:0061024(membrane organization);GO:0042981(regulation of apoptotic process);GO:0007166(cell surface receptor signaling pathway);GO:0051726(regulation of cell cycle);GO:0010803(regulation of tumor necrosis factor-mediated signaling pathway);GO:0000187(activation of MAPK activity);GO:0097194(execution phase of apoptosis);GO:0032483(regulation of Rab protein signal transduction);GO:0030295(protein kinase activator activity);GO:1902041(regulation of extrinsic apoptotic signaling pathway via death domain receptors);GO:0005123(death receptor binding);GO:2001236(regulation of extrinsic apoptotic signaling pathway) | NA | NA | NA | MAP kinase activating death domain [Source:HGNC Symbol;Acc:HGNC:6766] | 0.21 | 9.23 | 0.02 | -5.45128486752193 | 9.07e-12 | 3.24e-09 | down | yes |
| 47678 | NA | chr12 | - | 124776856 | 124863917 | ENST00000339570 | 12 | 3329 | ENSG00000073060 | SCARB1 | GO:0010595(positive regulation of endothelial cell migration);GO:0042632(cholesterol homeostasis);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0043231(intracellular membrane-bounded organelle);GO:0005765(lysosomal membrane);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0005901(caveola);GO:0006898(receptor-mediated endocytosis);GO:0046718(viral entry into host cell);GO:0070062(extracellular exosome);GO:0016032(viral process);GO:0009986(cell surface);GO:0005215(transporter activity);GO:0001530(lipopolysaccharide binding);GO:0030666(endocytic vesicle membrane);GO:0015914(phospholipid transport);GO:0051000(positive regulation of nitric-oxide synthase activity);GO:0001618(virus receptor activity);GO:0033344(cholesterol efflux);GO:0034384(high-density lipoprotein particle clearance);GO:0031663(lipopolysaccharide-mediated signaling pathway);GO:0034375(high-density lipoprotein particle remodeling);GO:0043691(reverse cholesterol transport);GO:0034185(apolipoprotein binding);GO:0034186(apolipoprotein A-I binding);GO:0030169(low-density lipoprotein particle binding);GO:0010867(positive regulation of triglyceride biosynthetic process);GO:0010899(regulation of phosphatidylcholine catabolic process);GO:0034383(low-density lipoprotein particle clearance);GO:0070508(cholesterol import);GO:0042060(wound healing);GO:0070328(triglyceride homeostasis);GO:0010886(positive regulation of cholesterol storage);GO:0044406(adhesion of symbiont to host);GO:0050764(regulation of phagocytosis);GO:0005044(scavenger receptor activity);GO:0015920(lipopolysaccharide transport);GO:0001786(phosphatidylserine binding);GO:0001875(lipopolysaccharide receptor activity);GO:0005545(1-phosphatidylinositol binding);GO:0070506(high-density lipoprotein particle receptor activity);GO:0032497(detection of lipopolysaccharide);GO:0035461(vitamin transmembrane transport);GO:0043654(recognition of apoptotic cell);GO:0001540(amyloid-beta binding);GO:0008035(high-density lipoprotein particle binding);GO:0042803(protein homodimerization activity);GO:0001935(endothelial cell proliferation);GO:0006702(androgen biosynthetic process);GO:0006707(cholesterol catabolic process);GO:0006869(lipid transport);GO:0006910(phagocytosis, recognition);GO:0030301(cholesterol transport);GO:0043534(blood vessel endothelial cell migration);GO:0050892(intestinal absorption);GO:0005737(cytoplasm);GO:0005887(integral component of plasma membrane);GO:0031528(microvillus membrane) | 04145(Phagosome);04913(Ovarian Steroidogenesis);04925(Aldosterone synthesis and secretion);04927(Cortisol synthesis and secretion);04976(Bile secretion);04975(Fat digestion and absorption);04979(Cholesterol metabolism);04977(Vitamin digestion and absorption);05160(Hepatitis C) | K13885 | NA | scavenger receptor class B member 1 [Source:HGNC Symbol;Acc:HGNC:1664] | 29.680565 | 0.74 | 40.1684729504048 | 5.33 | 1.03e-11 | 3.61e-09 | up | yes |
| 51060 | NA | chr13 | + | 102844844 | 102876001 | ENST00000355739 | 15 | 5082 | ENSG00000134899 | ERCC5 | GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0005654(nucleoplasm);GO:0016787(hydrolase activity);GO:0003677(DNA binding);GO:0003824(catalytic activity);GO:0042803(protein homodimerization activity);GO:0004519(endonuclease activity);GO:0006281(DNA repair);GO:0006974(cellular response to DNA damage stimulus);GO:0016788(hydrolase activity, acting on ester bonds);GO:0006289(nucleotide-excision repair);GO:0004518(nuclease activity);GO:0003697(single-stranded DNA binding);GO:0005515(protein binding);GO:0003690(double-stranded DNA binding);GO:0047485(protein N-terminus binding);GO:0043066(negative regulation of apoptotic process);GO:0009411(response to UV);GO:0006283(transcription-coupled nucleotide-excision repair);GO:0005662(DNA replication factor A complex);GO:0016591(DNA-directed RNA polymerase II, holoenzyme);GO:0006293(nucleotide-excision repair, preincision complex stabilization);GO:0006295(nucleotide-excision repair, DNA incision, 3'-to lesion);GO:0006296(nucleotide-excision repair, DNA incision, 5'-to lesion);GO:0033683(nucleotide-excision repair, DNA incision);GO:0006294(nucleotide-excision repair, preincision complex assembly);GO:0009650(UV protection);GO:0005675(holo TFIIH complex);GO:0010225(response to UV-C);GO:0004520(endodeoxyribonuclease activity);GO:0000405(bubble DNA binding) | 03420(Nucleotide excision repair) | K10846 | NA | ERCC excision repair 5, endonuclease [Source:HGNC Symbol;Acc:HGNC:3437] | 0.15 | 6.77 | 0.02 | -5.46407339795819 | 1.04e-11 | 3.61e-09 | down | yes |
| 126952 | NA | chr22 | - | 17137522 | 17143647 | ENST00000477157 | 3 | 3614 | ENSG00000069998 | HDHD5 | GO:0005739(mitochondrion);GO:0046474(glycerophospholipid biosynthetic process) | NA | NA | NA | haloacid dehalogenase like hydrolase domain containing 5 [Source:HGNC Symbol;Acc:HGNC:1843] | 7.03 | 0.17 | 42.1614752279936 | 5.40 | 1.20e-11 | 4.06e-09 | up | yes |
| 127316 | NA | chr22 | + | 19941773 | 19968958 | ENST00000403710 | 6 | 1548 | ENSG00000093010 | COMT | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0046872(metal ion binding);GO:0008168(methyltransferase activity);GO:0016740(transferase activity);GO:0032259(methylation);GO:0000287(magnesium ion binding);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0008171(O-methyltransferase activity);GO:0016206(catechol O-methyltransferase activity);GO:0006584(catecholamine metabolic process);GO:0007565(female pregnancy);GO:0007612(learning);GO:0007614(short-term memory);GO:0008210(estrogen metabolic process);GO:0009712(catechol-containing compound metabolic process);GO:0014070(response to organic cyclic compound);GO:0016036(cellular response to phosphate starvation);GO:0032496(response to lipopolysaccharide);GO:0032502(developmental process);GO:0035814(negative regulation of renal sodium excretion);GO:0042135(neurotransmitter catabolic process);GO:0042417(dopamine metabolic process);GO:0042420(dopamine catabolic process);GO:0042493(response to drug);GO:0045963(negative regulation of dopamine metabolic process);GO:0048265(response to pain);GO:0048609(multicellular organismal reproductive process);GO:0048662(negative regulation of smooth muscle cell proliferation);GO:0050668(positive regulation of homocysteine metabolic process);GO:0051930(regulation of sensory perception of pain);GO:0005739(mitochondrion);GO:0005829(cytosol);GO:0030424(axon);GO:0030425(dendrite);GO:0043197(dendritic spine);GO:0044297(cell body);GO:0045211(postsynaptic membrane);GO:0070062(extracellular exosome);GO:0102084(L-dopa O-methyltransferase activity);GO:0102938(-) | 00140(Steroid hormone biosynthesis);00350(Tyrosine metabolism);04728(Dopaminergic synapse) | K00545 | EC:2.1.1.6 | catechol-O-methyltransferase [Source:HGNC Symbol;Acc:HGNC:2228] | 19.418093 | 0.46 | 42.0656500886022 | 5.39 | 1.24e-11 | 4.07e-09 | up | yes |
| 129733 | NA | chr22 | + | 37675608 | 37679806 | ENST00000215909 | 4 | 560 | ENSG00000100097 | LGALS1 | GO:0005615(extracellular space);GO:0003723(RNA binding);GO:0005737(cytoplasm);GO:0030246(carbohydrate binding);GO:0005515(protein binding);GO:0004871(signal transducer activity);GO:0005576(extracellular region);GO:0006915(apoptotic process);GO:0005578(proteinaceous extracellular matrix);GO:0007165(signal transduction);GO:0070062(extracellular exosome);GO:0044267(cellular protein metabolic process);GO:0043687(post-translational protein modification);GO:0031012(extracellular matrix);GO:0043123(positive regulation of I-kappaB kinase/NF-kappaB signaling);GO:0042981(regulation of apoptotic process);GO:0046598(positive regulation of viral entry into host cell);GO:0005788(endoplasmic reticulum lumen);GO:0005622(intracellular);GO:0030395(lactose binding);GO:0042803(protein homodimerization activity);GO:0043236(laminin binding);GO:0002317(plasma cell differentiation);GO:0010812(negative regulation of cell-substrate adhesion);GO:0010977(negative regulation of neuron projection development);GO:0031295(T cell costimulation);GO:0034120(positive regulation of erythrocyte aggregation);GO:0035900(response to isolation stress);GO:0042493(response to drug);GO:0045445(myoblast differentiation);GO:0048678(response to axon injury);GO:0071333(cellular response to glucose stimulus);GO:0071407(cellular response to organic cyclic compound);GO:0005634(nucleus);GO:0005829(cytosol);GO:0009986(cell surface) | NA | NA | NA | galectin 1 [Source:HGNC Symbol;Acc:HGNC:6561] | 4.01 | 166.562683 | 0.02 | -5.37641810736717 | 1.24e-11 | 4.07e-09 | down | yes |
| 68187 | NA | chr16 | - | 3658038 | 3678304 | ENST00000575671 | 13 | 2309 | ENSG00000126602 | TRAP1 | GO:0005524(ATP binding);GO:0006457(protein folding);GO:0051082(unfolded protein binding);GO:0006950(response to stress);GO:0003723(RNA binding);GO:0016020(membrane);GO:0000166(nucleotide binding);GO:0005654(nucleoplasm);GO:0005739(mitochondrion);GO:0005743(mitochondrial inner membrane);GO:0005759(mitochondrial matrix);GO:0005515(protein binding);GO:0019901(protein kinase binding);GO:1903751(negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide);GO:0005758(mitochondrial intermembrane space);GO:0070062(extracellular exosome);GO:0005164(tumor necrosis factor receptor binding);GO:1903427(negative regulation of reactive oxygen species biosynthetic process);GO:0009386(translational attenuation);GO:0061077(chaperone-mediated protein folding);GO:1901856(negative regulation of cellular respiration);GO:0005811(lipid particle) | NA | NA | NA | TNF receptor associated protein 1 [Source:HGNC Symbol;Acc:HGNC:16264] | 15.59778 | 0.38 | 40.8588388151346 | 5.35 | 1.33e-11 | 4.28e-09 | up | yes |
| 43423 | NA | chr12 | - | 64713445 | 64752871 | ENST00000418919 | 13 | 4877 | ENSG00000135677 | GNS | GO:0003824(catalytic activity);GO:0008152(metabolic process);GO:0005764(lysosome);GO:0008484(sulfuric ester hydrolase activity);GO:0008449(N-acetylglucosamine-6-sulfatase activity);GO:0030203(glycosaminoglycan metabolic process);GO:0046872(metal ion binding);GO:0016787(hydrolase activity);GO:0005515(protein binding);GO:0005576(extracellular region);GO:0070062(extracellular exosome);GO:0043312(neutrophil degranulation);GO:1904813(ficolin-1-rich granule lumen);GO:0043202(lysosomal lumen);GO:0035578(azurophil granule lumen);GO:0042340(keratan sulfate catabolic process);GO:0006027(glycosaminoglycan catabolic process) | 00531(Glycosaminoglycan degradation);04142(Lysosome) | NA | NA | glucosamine (N-acetyl)-6-sulfatase [Source:HGNC Symbol;Acc:HGNC:4422] | 49.84037 | 1.29 | 38.560588183867 | 5.27 | 1.39e-11 | 4.39e-09 | up | yes |
| 55435 | NA | chr14 | - | 67338071 | 67360249 | ENST00000216442 | 9 | 1845 | ENSG00000100554 | ATP6V1D | GO:0006810(transport);GO:0016020(membrane);GO:0005765(lysosomal membrane);GO:0055085(transmembrane transport);GO:0006811(ion transport);GO:0005515(protein binding);GO:0015992(proton transport);GO:0042626(ATPase activity, coupled to transmembrane movement of substances);GO:0005813(centrosome);GO:0005886(plasma membrane);GO:0060271(cilium assembly);GO:0005929(cilium);GO:0005829(cytosol);GO:0070062(extracellular exosome);GO:0034220(ion transmembrane transport);GO:0030030(cell projection organization);GO:0043312(neutrophil degranulation);GO:0035579(specific granule membrane);GO:0008286(insulin receptor signaling pathway);GO:0016241(regulation of macroautophagy);GO:0033572(transferrin transport);GO:0090383(phagosome acidification);GO:0061512(protein localization to cilium);GO:0033176(proton-transporting V-type ATPase complex) | 00190(Oxidative phosphorylation);04150(mTOR signaling pathway);04145(Phagosome);04966(Collecting duct acid secretion);04721(Synaptic vesicle cycle);05323(Rheumatoid arthritis);05110(Vibrio cholerae infection);05120(Epithelial cell signaling in Helicobacter pylori infection) | K02149 | NA | ATPase H+ transporting V1 subunit D [Source:HGNC Symbol;Acc:HGNC:13527] | 17.901655 | 0.44 | 40.4617524873766 | 5.34 | 1.51e-11 | 4.70e-09 | up | yes |
| 53439 | NA | chr14 | - | 31100112 | 31127868 | ENST00000554882 | 18 | 3748 | ENSG00000092148 | HECTD1 | GO:0016740(transferase activity);GO:0016567(protein ubiquitination);GO:0004842(ubiquitin-protein transferase activity);GO:0046872(metal ion binding);GO:0005737(cytoplasm);GO:0061630(ubiquitin protein ligase activity);GO:0005515(protein binding) | NA | NA | NA | HECT domain E3 ubiquitin protein ligase 1 [Source:HGNC Symbol;Acc:HGNC:20157] | 0.17 | 7.03 | 0.02 | -5.38253724543935 | 2.09e-11 | 6.36e-09 | down | yes |
| 81579 | NA | chr17 | - | 36495638 | 36535415 | ENST00000610992 | 27 | 5743 | ENSG00000278259 | MYO19 | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0003774(motor activity);GO:0016459(myosin complex);GO:0003779(actin binding);GO:0005739(mitochondrion);GO:0005856(cytoskeleton);GO:0005741(mitochondrial outer membrane);GO:0016887(ATPase activity);GO:0032465(regulation of cytokinesis);GO:0090140(regulation of mitochondrial fission);GO:0032027(myosin light chain binding);GO:0060002(plus-end directed microfilament motor activity) | NA | NA | NA | myosin XIX [Source:HGNC Symbol;Acc:HGNC:26234] | 8.30 | 0.22 | 37.2479917111832 | 5.22 | 2.65e-11 | 7.93e-09 | up | yes |
| 172331 | NA | chr7 | - | 51016213 | 51316818 | ENST00000395542 | 14 | 5339 | ENSG00000106078 | COBL | GO:0003779(actin binding);GO:0048471(perinuclear region of cytoplasm);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0005886(plasma membrane);GO:0005856(cytoskeleton);GO:0042995(cell projection);GO:0048565(digestive tract development);GO:0001726(ruffle);GO:0000578(embryonic axis specification);GO:0001843(neural tube closure);GO:0030424(axon);GO:0030041(actin filament polymerization);GO:0005884(actin filament);GO:0003785(actin monomer binding);GO:0001757(somite specification);GO:0001889(liver development);GO:0030903(notochord development);GO:0033504(floor plate development);GO:0048669(collateral sprouting in absence of injury);GO:0051639(actin filament network formation);GO:1900006(positive regulation of dendrite development);GO:0005938(cell cortex);GO:0030425(dendrite);GO:0043025(neuronal cell body);GO:0044294(dendritic growth cone);GO:0044295(axonal growth cone);GO:1900029(positive regulation of ruffle assembly) | NA | NA | NA | cordon-bleu WH2 repeat protein [Source:HGNC Symbol;Acc:HGNC:22199] | 6.69 | 0.18 | 37.3352287946429 | 5.22 | 3.02e-11 | 8.89e-09 | up | yes |
| 120358 | NA | chr20 | - | 2658395 | 2664188 | ENST00000488299 | 11 | 1620 | ENSG00000101365 | IDH3B | GO:0000287(magnesium ion binding);GO:0055114(oxidation-reduction process);GO:0016616(oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor);GO:0051287(NAD binding);GO:0046872(metal ion binding);GO:0016491(oxidoreductase activity);GO:0006099(tricarboxylic acid cycle);GO:0005739(mitochondrion);GO:0004449(isocitrate dehydrogenase (NAD+) activity);GO:0006102(isocitrate metabolic process);GO:0006103(2-oxoglutarate metabolic process);GO:0006734(NADH metabolic process);GO:0005634(nucleus);GO:0005759(mitochondrial matrix);GO:0009055(electron carrier activity) | 00020(Citrate cycle (TCA cycle)) | K00030 | EC:1.1.1.41 | isocitrate dehydrogenase 3 (NAD(+)) beta [Source:HGNC Symbol;Acc:HGNC:5385] | 39.357193 | 1.10 | 35.7041254087988 | 5.16 | 3.64e-11 | 1.05e-08 | up | yes |
| 79388 | NA | chr17 | - | 18244836 | 18258726 | ENST00000578558 | 17 | 2377 | ENSG00000177731 | FLII | GO:0051015(actin filament binding);GO:0030036(actin cytoskeleton organization);GO:0051014(actin filament severing);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0005815(microtubule organizing center);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003779(actin binding);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0007275(multicellular organism development);GO:0030054(cell junction);GO:0005925(focal adhesion);GO:0005903(brush border) | NA | NA | NA | FLII, actin remodeling protein [Source:HGNC Symbol;Acc:HGNC:3750] | 0.87 | 31.882957 | 0.03 | -5.20224461737225 | 3.89e-11 | 1.11e-08 | down | yes |
| 133690 | NA | chr3 | + | 41199445 | 41239736 | ENST00000453024 | 17 | 2841 | ENSG00000168036 | CTNNB1 | GO:0048471(perinuclear region of cytoplasm);GO:0016525(negative regulation of angiogenesis);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0005737(cytoplasm);GO:0005634(nucleus);GO:0061154(endothelial tube morphogenesis);GO:0016020(membrane);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0004871(signal transducer activity);GO:0007155(cell adhesion);GO:0005886(plasma membrane);GO:0005829(cytosol);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0034333(adherens junction assembly);GO:0016055(Wnt signaling pathway);GO:0030054(cell junction);GO:0045202(synapse);GO:0007399(nervous system development);GO:0008134(transcription factor binding);GO:0003713(transcription coactivator activity);GO:0045893(positive regulation of transcription, DNA-templated);GO:0005925(focal adhesion);GO:0000922(spindle pole);GO:0005813(centrosome);GO:0005815(microtubule organizing center);GO:0005654(nucleoplasm);GO:0045296(cadherin binding);GO:0070062(extracellular exosome);GO:0005622(intracellular);GO:0008285(negative regulation of cell proliferation);GO:0043161(proteasome-mediated ubiquitin-dependent protein catabolic process);GO:0016323(basolateral plasma membrane);GO:0005912(adherens junction);GO:0043065(positive regulation of apoptotic process);GO:0043234(protein complex);GO:0008022(protein C-terminus binding);GO:0005913(cell-cell adherens junction);GO:0005667(transcription factor complex);GO:0044325(ion channel binding);GO:0019899(enzyme binding);GO:0019903(protein phosphatase binding);GO:0044212(transcription regulatory region DNA binding);GO:0045892(negative regulation of transcription, DNA-templated);GO:1904837(beta-catenin-TCF complex assembly);GO:0030521(androgen receptor signaling pathway);GO:0001102(RNA polymerase II activating transcription factor binding);GO:0032355(response to estradiol);GO:1904886(beta-catenin destruction complex disassembly);GO:0001569(branching involved in blood vessel morphogenesis);GO:0032481(positive regulation of type I interferon production);GO:1904948(midbrain dopaminergic neuron differentiation);GO:0005911(cell-cell junction);GO:0016342(catenin complex);GO:0070411(I-SMAD binding);GO:0046332(SMAD binding);GO:0060070(canonical Wnt signaling pathway);GO:0016328(lateral plasma membrane);GO:0030877(beta-catenin destruction complex);GO:0071944(cell periphery);GO:0050681(androgen receptor binding);GO:0007223(Wnt signaling pathway, calcium modulating pathway);GO:0051091(positive regulation of sequence-specific DNA binding transcription factor activity);GO:0071363(cellular response to growth factor stimulus);GO:0045765(regulation of angiogenesis);GO:0019900(kinase binding);GO:0051149(positive regulation of muscle cell differentiation);GO:0032993(protein-DNA complex);GO:0035635(entry of bacterium into host cell);GO:0019827(stem cell population maintenance);GO:0005938(cell cortex);GO:1990909(Wnt signalosome);GO:0035315(hair cell differentiation);GO:0042493(response to drug);GO:0016337(single organismal cell-cell adhesion);GO:0000978(RNA polymerase II core promoter proximal region sequence-specific DNA binding);GO:0090279(regulation of calcium ion import);GO:2000008(regulation of protein localization to cell surface);GO:0044336(canonical Wnt signaling pathway involved in negative regulation of apoptotic process);GO:1904954(canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation);GO:0005719(nuclear euchromatin);GO:0060548(negative regulation of cell death);GO:0071681(cellular response to indole-3-methanol);GO:0001837(epithelial to mesenchymal transition);GO:0045294(alpha-catenin binding);GO:0034394(protein localization to cell surface);GO:0001085(RNA polymerase II transcription factor binding);GO:0033234(negative regulation of protein sumoylation);GO:0072182(regulation of nephron tubule epithelial cell differentiation);GO:0030331(estrogen receptor binding);GO:0035257(nuclear hormone receptor binding);GO:0002052(positive regulation of neuroblast proliferation);GO:0010718(positive regulation of epithelial to mesenchymal transition);GO:0010909(positive regulation of heparan sulfate proteoglycan biosynthetic process);GO:0030997(regulation of centriole-centriole cohesion);GO:0035411(catenin import into nucleus);GO:0036023(embryonic skeletal limb joint morphogenesis);GO:0043525(positive regulation of neuron apoptotic process);GO:0044334(canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition);GO:0045976(negative regulation of mitotic cell cycle, embryonic);GO:0048145(regulation of fibroblast proliferation);GO:0048660(regulation of smooth muscle cell proliferation);GO:0050767(regulation of neurogenesis);GO:0051571(positive regulation of histone H3-K4 methylation);GO:0061324(canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation);GO:0061549(sympathetic ganglion development);GO:0070602(regulation of centromeric sister chromatid cohesion);GO:1904798(positive regulation of core promoter binding);GO:2000144(positive regulation of DNA-templated transcription, initiation);GO:0070369(beta-catenin-TCF7L2 complex);GO:1990907(beta-catenin-TCF complex);GO:0007165(signal transduction);GO:0000979(RNA polymerase II core promoter sequence-specific DNA binding);GO:0003677(DNA binding);GO:0003682(chromatin binding);GO:0003690(double-stranded DNA binding);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0019901(protein kinase binding);GO:0046982(protein heterodimerization activity);GO:0070491(repressing transcription factor binding);GO:0097718(disordered domain specific binding);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0000578(embryonic axis specification);GO:0000904(cell morphogenesis involved in differentiation);GO:0001501(skeletal system development);GO:0001570(vasculogenesis);GO:0001658(branching involved in ureteric bud morphogenesis);GO:0001701(in utero embryonic development);GO:0001702(gastrulation with mouth forming second);GO:0001706(endoderm formation);GO:0001708(cell fate specification);GO:0001709(cell fate determination);GO:0001711(endodermal cell fate commitment);GO:0001764(neuron migration);GO:0001822(kidney development);GO:0001840(neural plate development);GO:0001944(vasculature development);GO:0002053(positive regulation of mesenchymal cell proliferation);GO:0002089(lens morphogenesis in camera-type eye);GO:0003266(regulation of secondary heart field cardioblast proliferation);GO:0003338(metanephros morphogenesis);GO:0003340(negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis);GO:0006357(regulation of transcription from RNA polymerase II promoter);GO:0007160(cell-matrix adhesion);GO:0007268(chemical synaptic transmission);GO:0007398(ectoderm development);GO:0007403(glial cell fate determination);GO:0007507(heart development);GO:0008283(cell proliferation);GO:0008284(positive regulation of cell proliferation);GO:0009948(anterior/posterior axis specification);GO:0009950(dorsal/ventral axis specification);GO:0009953(dorsal/ventral pattern formation);GO:0009954(proximal/distal pattern formation);GO:0009987(cellular process);GO:0010468(regulation of gene expression);GO:0010628(positive regulation of gene expression);GO:0010629(negative regulation of gene expression);GO:0016331(morphogenesis of embryonic epithelium);GO:0021819(layer formation in cerebral cortex);GO:0022009(central nervous system vasculogenesis);GO:0022405(hair cycle process);GO:0030097(hemopoiesis);GO:0030154(cell differentiation);GO:0030182(neuron differentiation);GO:0030217(T cell differentiation);GO:0030316(osteoclast differentiation);GO:0030324(lung development);GO:0030539(male genitalia development);GO:0030856(regulation of epithelial cell differentiation);GO:0030858(positive regulation of epithelial cell differentiation);GO:0030900(forebrain development);GO:0030901(midbrain development);GO:0030902(hindbrain development);GO:0031016(pancreas development);GO:0031069(hair follicle morphogenesis);GO:0031641(regulation of myelination);GO:0032212(positive regulation of telomere maintenance via telomerase);GO:0032331(negative regulation of chondrocyte differentiation);GO:0033077(T cell differentiation in thymus);GO:0034332(adherens junction organization);GO:0034613(cellular protein localization);GO:0035050(embryonic heart tube development);GO:0035112(genitalia morphogenesis);GO:0035115(embryonic forelimb morphogenesis);GO:0035116(embryonic hindlimb morphogenesis);GO:0042127(regulation of cell proliferation);GO:0042129(regulation of T cell proliferation);GO:0042475(odontogenesis of dentin-containing tooth);GO:0042733(embryonic digit morphogenesis);GO:0042981(regulation of apoptotic process);GO:0043123(positive regulation of I-kappaB kinase/NF-kappaB signaling);GO:0043410(positive regulation of MAPK cascade);GO:0043588(skin development);GO:0045453(bone resorption);GO:0045595(regulation of cell differentiation);GO:0045596(negative regulation of cell differentiation);GO:0045603(positive regulation of endothelial cell differentiation);GO:0045667(regulation of osteoblast differentiation);GO:0045669(positive regulation of osteoblast differentiation);GO:0045670(regulation of osteoclast differentiation);GO:0045671(negative regulation of osteoclast differentiation);GO:0045743(positive regulation of fibroblast growth factor receptor signaling pathway);GO:0048096(chromatin-mediated maintenance of transcription);GO:0048469(cell maturation);GO:0048489(synaptic vesicle transport);GO:0048513(animal organ development);GO:0048538(thymus development);GO:0048599(oocyte development);GO:0048617(embryonic foregut morphogenesis);GO:0048643(positive regulation of skeletal muscle tissue development);GO:0048715(negative regulation of oligodendrocyte differentiation);GO:0050808(synapse organization);GO:0051145(smooth muscle cell differentiation);GO:0051884(regulation of timing of anagen);GO:0051973(positive regulation of telomerase activity);GO:0060066(oviduct development);GO:0060173(limb development);GO:0060439(trachea morphogenesis);GO:0060440(trachea formation);GO:0060441(epithelial tube branching involved in lung morphogenesis);GO:0060479(lung cell differentiation);GO:0060484(lung-associated mesenchyme development);GO:0060492(lung induction);GO:0060742(epithelial cell differentiation involved in prostate gland development);GO:0060769(positive regulation of epithelial cell proliferation involved in prostate gland development);GO:0060789(hair follicle placode formation);GO:0060916(mesenchymal cell proliferation involved in lung development);GO:0061047(positive regulation of branching involved in lung morphogenesis);GO:0061198(fungiform papilla formation);GO:0061550(cranial ganglion development);GO:0072001(renal system development);GO:0072033(renal vesicle formation);GO:0072053(renal inner medulla development);GO:0072054(renal outer medulla development);GO:0072079(nephron tubule formation);GO:1901215(negative regulation of neuron death);GO:1903204(negative regulation of oxidative stress-induced neuron death);GO:1904501(positive regulation of chromatin-mediated maintenance of transcription);GO:1904793(regulation of euchromatin binding);GO:1904796(regulation of core promoter binding);GO:1904888(cranial skeletal system development);GO:1990403(embryonic brain development);GO:1990791(dorsal root ganglion development);GO:2000017(positive regulation of determination of dorsal identity);GO:2001234(negative regulation of apoptotic signaling pathway);GO:0005916(fascia adherens);GO:0005923(bicellular tight junction);GO:0014704(intercalated disc);GO:0016600(flotillin complex);GO:0030018(Z disc);GO:0030027(lamellipodium);GO:0031253(cell projection membrane);GO:0031528(microvillus membrane);GO:0034750(Scrib-APC-beta-catenin complex);GO:0043296(apical junction complex);GO:0044798(nuclear transcription factor complex);GO:0045177(apical part of cell);GO:0071664(catenin-TCF7L2 complex) | 04015(Rap1 signaling pathway);04310(Wnt signaling pathway);04390(Hippo signaling pathway);04510(Focal adhesion);04520(Adherens junction);04550(Signaling pathways regulating pluripotency of stem cells);04670(Leukocyte transendothelial migration);04919(Thyroid hormone signaling pathway);04916(Melanogenesis);05200(Pathways in cancer);05205(Proteoglycans in cancer);05210(Colorectal cancer);05225(Hepatocellular carcinoma);05226(Gastric cancer);05216(Thyroid cancer);05217(Basal cell carcinoma);05215(Prostate cancer);05213(Endometrial cancer);05224(Breast cancer);05418(Fluid shear stress and atherosclerosis);05412(Arrhythmogenic right ventricular cardiomyopathy (ARVC));05130(Pathogenic Escherichia coli infection);05100(Bacterial invasion of epithelial cells);05166(HTLV-I infection);05167(Kaposi's sarcoma-associated herpesvirus infection);05165(Human papillomavirus infection) | K02105 | NA | catenin beta 1 [Source:HGNC Symbol;Acc:HGNC:2514] | 0.31 | 11.80121 | 0.03 | -5.24639140758612 | 4.00e-11 | 1.12e-08 | down | yes |
| 172016 | NA | chr7 | + | 44796680 | 44803117 | ENST00000468812 | 5 | 2238 | ENSG00000196262 | PPIA | GO:0016853(isomerase activity);GO:0003755(peptidyl-prolyl cis-trans isomerase activity);GO:0000413(protein peptidyl-prolyl isomerization);GO:0006457(protein folding);GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0005829(cytosol);GO:0005515(protein binding);GO:0005576(extracellular region);GO:0005615(extracellular space);GO:0005925(focal adhesion);GO:0050900(leukocyte migration);GO:0070062(extracellular exosome);GO:0019058(viral life cycle);GO:0016032(viral process);GO:0043312(neutrophil degranulation);GO:0035722(interleukin-12-mediated signaling pathway);GO:0034774(secretory granule lumen);GO:1904813(ficolin-1-rich granule lumen);GO:0031982(vesicle);GO:0050714(positive regulation of protein secretion);GO:0051082(unfolded protein binding);GO:0045070(positive regulation of viral genome replication);GO:0019076(viral release from host cell);GO:0075713(establishment of integrated proviral latency);GO:0019064(fusion of virus membrane with host plasma membrane);GO:0030260(entry into host cell);GO:0019068(virion assembly);GO:0046790(virion binding);GO:0016018(cyclosporin A binding);GO:0006278(RNA-dependent DNA biosynthetic process);GO:0019061(uncoating of virus);GO:0034389(lipid particle organization);GO:0045069(regulation of viral genome replication) | 04217(Necroptosis) | K03767 | EC:5.2.1.8 | peptidylprolyl isomerase A [Source:HGNC Symbol;Acc:HGNC:9253] | 70.532005 | 2.05 | 34.4686563079562 | 5.11 | 4.15e-11 | 1.14e-08 | up | yes |
| 192193 | NA | chr9 | - | 129636892 | 129642165 | ENST00000277458 | 6 | 2330 | ENSG00000148331 | ASB6 | GO:0005737(cytoplasm);GO:0016567(protein ubiquitination);GO:0035556(intracellular signal transduction);GO:0005515(protein binding);GO:0005829(cytosol);GO:0043687(post-translational protein modification);GO:0005622(intracellular) | NA | NA | NA | ankyrin repeat and SOCS box containing 6 [Source:HGNC Symbol;Acc:HGNC:17181] | 15.167649 | 0.42 | 35.8281893684569 | 5.16 | 4.34e-11 | 1.18e-08 | up | yes |
| 160437 | NA | chr5 | - | 179798165 | 179800597 | ENST00000520875 | 8 | 964 | ENSG00000161013 | MGAT4B | GO:0016020(membrane);GO:0005975(carbohydrate metabolic process);GO:0016758(transferase activity, transferring hexosyl groups);GO:0016021(integral component of membrane);GO:0046872(metal ion binding);GO:0016740(transferase activity);GO:0016757(transferase activity, transferring glycosyl groups);GO:0005794(Golgi apparatus);GO:0000139(Golgi membrane);GO:0006486(protein glycosylation);GO:0006491(N-glycan processing);GO:0008454(alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity);GO:0008375(acetylglucosaminyltransferase activity);GO:0006487(protein N-linked glycosylation) | 00510(N-Glycan biosynthesis) | NA | NA | mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B [Source:HGNC Symbol;Acc:HGNC:7048] | 0.99 | 37.132198 | 0.03 | -5.22347700724267 | 4.43e-11 | 1.19e-08 | down | yes |
| 40896 | NA | chr12 | + | 49750620 | 49762948 | ENST00000552370 | 10 | 882 | ENSG00000139644 | TMBIM6 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0043066(negative regulation of apoptotic process);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0006915(apoptotic process);GO:0006914(autophagy);GO:0006986(response to unfolded protein);GO:0005887(integral component of plasma membrane);GO:0005737(cytoplasm);GO:0005634(nucleus);GO:0005515(protein binding);GO:0031625(ubiquitin protein ligase binding);GO:1902236(negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway);GO:0019899(enzyme binding);GO:0032091(negative regulation of protein binding);GO:2001234(negative regulation of apoptotic signaling pathway);GO:0031966(mitochondrial membrane);GO:0060698(endoribonuclease inhibitor activity);GO:0010523(negative regulation of calcium ion transport into cytosol);GO:0060702(negative regulation of endoribonuclease activity);GO:1902065(response to L-glutamate);GO:1903298(negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway);GO:1904721(negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response);GO:0032469(endoplasmic reticulum calcium ion homeostasis);GO:0034976(response to endoplasmic reticulum stress);GO:0051025(negative regulation of immunoglobulin secretion);GO:0070059(intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress);GO:1990441(negative regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress) | NA | NA | NA | transmembrane BAX inhibitor motif containing 6 [Source:HGNC Symbol;Acc:HGNC:11723] | 0.83 | 31.078743 | 0.03 | -5.23114533428571 | 4.73e-11 | 1.25e-08 | down | yes |
| 79642 | NA | chr17 | - | 18975710 | 19003780 | ENST00000399096 | 3 | 4393 | ENSG00000188522 | FAM83G | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0005829(cytosol);GO:0005515(protein binding);GO:0030509(BMP signaling pathway) | NA | NA | NA | family with sequence similarity 83 member G [Source:HGNC Symbol;Acc:HGNC:32554] | 0.26 | 9.37 | 0.03 | -5.19284699176129 | 4.99e-11 | 1.30e-08 | down | yes |
| 177084 | NA | chr7 | - | 132123332 | 132576564 | ENST00000321063 | 32 | 13061 | ENSG00000221866 | PLXNA4 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0007165(signal transduction);GO:0005886(plasma membrane);GO:0017154(semaphorin receptor activity);GO:0007399(nervous system development);GO:0007411(axon guidance);GO:0008045(motor neuron axon guidance);GO:0021602(cranial nerve morphogenesis);GO:0021610(facial nerve morphogenesis);GO:0021612(facial nerve structural organization);GO:0021615(glossopharyngeal nerve morphogenesis);GO:0021636(trigeminal nerve morphogenesis);GO:0021637(trigeminal nerve structural organization);GO:0021644(vagus nerve morphogenesis);GO:0021784(postganglionic parasympathetic fiber development);GO:0021785(branchiomotor neuron axon guidance);GO:0021793(chemorepulsion of branchiomotor axon);GO:0021960(anterior commissure morphogenesis);GO:0048485(sympathetic nervous system development);GO:0048812(neuron projection morphogenesis);GO:0048841(regulation of axon extension involved in axon guidance);GO:0050923(regulation of negative chemotaxis);GO:0071526(semaphorin-plexin signaling pathway);GO:1902287(semaphorin-plexin signaling pathway involved in axon guidance);GO:0002116(semaphorin receptor complex) | 04360(Axon guidance) | K06820 | NA | plexin A4 [Source:HGNC Symbol;Acc:HGNC:9102] | 3.34 | 0.10 | 34.8003127769379 | 5.12 | 5.07e-11 | 1.30e-08 | up | yes |
| 110587 | NA | chr2 | + | 85539165 | 85545280 | ENST00000306434 | 9 | 2819 | ENSG00000168906 | MAT2A | GO:0046872(metal ion binding);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0016740(transferase activity);GO:0006730(one-carbon metabolic process);GO:0005515(protein binding);GO:0042802(identical protein binding);GO:0004478(methionine adenosyltransferase activity);GO:0006556(S-adenosylmethionine biosynthetic process);GO:1990830(cellular response to leukemia inhibitory factor);GO:0005829(cytosol);GO:0051291(protein heterooligomerization);GO:0034214(protein hexamerization);GO:0032259(methylation);GO:0048269(methionine adenosyltransferase complex) | 00270(Cysteine and methionine metabolism) | K00789 | EC:2.5.1.6 | methionine adenosyltransferase 2A [Source:HGNC Symbol;Acc:HGNC:6904] | 37.495411 | 1.12 | 33.5358949545108 | 5.07 | 5.67e-11 | 1.42e-08 | up | yes |
| 17866 | NA | chr10 | - | 3775996 | 3785275 | ENST00000497571 | 4 | 4656 | ENSG00000067082 | KLF6 | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0043231(intracellular membrane-bounded organelle);GO:0046872(metal ion binding);GO:0005730(nucleolus);GO:0005829(cytosol);GO:0006366(transcription from RNA polymerase II promoter);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0001650(fibrillar center);GO:0005515(protein binding);GO:0045893(positive regulation of transcription, DNA-templated);GO:0000978(RNA polymerase II core promoter proximal region sequence-specific DNA binding);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0001077(transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding);GO:0030183(B cell differentiation) | NA | NA | NA | Kruppel like factor 6 [Source:HGNC Symbol;Acc:HGNC:2235] | 0.29 | 10.366919 | 0.03 | -5.16372095984732 | 5.68e-11 | 1.42e-08 | down | yes |
| 192713 | NA | chr9 | + | 135499984 | 135504673 | ENST00000371785 | 5 | 1640 | ENSG00000122140 | MRPS2 | GO:0005739(mitochondrion);GO:0030529(intracellular ribonucleoprotein complex);GO:0005622(intracellular);GO:0003735(structural constituent of ribosome);GO:0006412(translation);GO:0005840(ribosome);GO:0015935(small ribosomal subunit);GO:0032543(mitochondrial translation);GO:0005743(mitochondrial inner membrane);GO:0070125(mitochondrial translational elongation);GO:0070126(mitochondrial translational termination);GO:0005763(mitochondrial small ribosomal subunit) | 03010(Ribosome) | K02967 | NA | mitochondrial ribosomal protein S2 [Source:HGNC Symbol;Acc:HGNC:14495] | 2.49 | 85.386536 | 0.03 | -5.10157485468551 | 6.93e-11 | 1.71e-08 | down | yes |
| 117700 | NA | chr2 | + | 216633429 | 216664433 | ENST00000490362 | 2 | 4414 | ENSG00000115457 | IGFBP2 | GO:0005576(extracellular region);GO:0005520(insulin-like growth factor binding);GO:0032868(response to insulin);GO:0005615(extracellular space);GO:0042104(positive regulation of activated T cell proliferation);GO:0005515(protein binding);GO:0001558(regulation of cell growth);GO:0070062(extracellular exosome);GO:0044267(cellular protein metabolic process);GO:0040008(regulation of growth);GO:0019838(growth factor binding);GO:0090090(negative regulation of canonical Wnt signaling pathway);GO:0031994(insulin-like growth factor I binding);GO:0005102(receptor binding);GO:0031995(insulin-like growth factor II binding);GO:0043567(regulation of insulin-like growth factor receptor signaling pathway);GO:0007165(signal transduction);GO:0007565(female pregnancy);GO:0007568(aging);GO:0007584(response to nutrient);GO:0009612(response to mechanical stimulus);GO:0010226(response to lithium ion);GO:0032355(response to estradiol);GO:0032526(response to retinoic acid);GO:0032870(cellular response to hormone stimulus);GO:0042493(response to drug);GO:0043627(response to estrogen);GO:0048545(response to steroid hormone);GO:0051384(response to glucocorticoid);GO:0016324(apical plasma membrane);GO:0031410(cytoplasmic vesicle) | NA | NA | NA | insulin like growth factor binding protein 2 [Source:HGNC Symbol;Acc:HGNC:5471] | 0.21 | 7.63 | 0.03 | -5.15146562523445 | 7.25e-11 | 1.76e-08 | down | yes |
| 168433 | NA | chr6 | + | 149977922 | 149978416 | ENST00000433415 | 1 | 495 | ENSG00000231120 | BTF3P10 | NA | NA | NA | NA | basic transcription factor 3 pseudogene 10 [Source:HGNC Symbol;Acc:HGNC:38570] | 175.663574 | 5.37 | 32.6839666506035 | 5.03 | 7.45e-11 | 1.79e-08 | up | yes |
| 76317 | NA | chr16 | + | 89921851 | 89936091 | ENST00000554444 | 4 | 1978 | ENSG00000258947 | TUBB3 | GO:0005525(GTP binding);GO:0000166(nucleotide binding);GO:0003924(GTPase activity);GO:0015630(microtubule cytoskeleton);GO:0005200(structural constituent of cytoskeleton);GO:0007010(cytoskeleton organization);GO:0005874(microtubule);GO:0007017(microtubule-based process);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0070062(extracellular exosome);GO:0007411(axon guidance);GO:0030425(dendrite) | 04145(Phagosome);04540(Gap junction);05130(Pathogenic Escherichia coli infection) | K07375 | NA | tubulin beta 3 class III [Source:HGNC Symbol;Acc:HGNC:20772] | 18.970385 | 0.57 | 33.5105422697676 | 5.07 | 7.63e-11 | 1.81e-08 | up | yes |
| 28927 | NA | chr11 | - | 57325986 | 57335851 | ENST00000293880 | 11 | 6296 | ENSG00000149136 | SSRP1 | GO:0005634(nucleus);GO:0003723(RNA binding);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0005694(chromosome);GO:0005515(protein binding);GO:0006281(DNA repair);GO:0006974(cellular response to DNA damage stimulus);GO:0006260(DNA replication);GO:0003682(chromatin binding);GO:0005730(nucleolus);GO:0005654(nucleoplasm);GO:0006366(transcription from RNA polymerase II promoter);GO:0006368(transcription elongation from RNA polymerase II promoter);GO:1901796(regulation of signal transduction by p53 class mediator) | NA | NA | NA | structure specific recognition protein 1 [Source:HGNC Symbol;Acc:HGNC:11327] | 0.90 | 29.662037 | 0.03 | -5.04036563592708 | 9.82e-11 | 2.30e-08 | down | yes |
| 17356 | NA | chr1 | - | 244840638 | 244854748 | ENST00000366527 | 2 | 12852 | ENSG00000153187 | HNRNPU | GO:0005634(nucleus);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0003723(RNA binding);GO:0006397(mRNA processing);GO:0008380(RNA splicing);GO:0005737(cytoplasm);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0005654(nucleoplasm);GO:0003677(DNA binding);GO:0030529(intracellular ribonucleoprotein complex);GO:0005515(protein binding);GO:0005681(spliceosomal complex);GO:0048511(rhythmic process);GO:0001047(core promoter binding);GO:0031012(extracellular matrix);GO:0000398(mRNA splicing, via spliceosome);GO:0009986(cell surface);GO:0032922(circadian regulation of gene expression);GO:0036464(cytoplasmic ribonucleoprotein granule);GO:0070034(telomerase RNA binding);GO:0005697(telomerase holoenzyme complex);GO:0016070(RNA metabolic process);GO:0006396(RNA processing);GO:0071013(catalytic step 2 spliceosome);GO:0001649(osteoblast differentiation);GO:0070934(CRD-mediated mRNA stabilization);GO:0070937(CRD-mediated mRNA stability complex);GO:0032211(negative regulation of telomere maintenance via telomerase);GO:0034046(poly(G) binding);GO:0035326(enhancer binding);GO:0043021(ribonucleoprotein complex binding);GO:0010628(positive regulation of gene expression);GO:0055013(cardiac muscle cell development);GO:0071549(cellular response to dexamethasone stimulus);GO:2000373(positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity) | 03040(Spliceosome) | NA | NA | heterogeneous nuclear ribonucleoprotein U [Source:HGNC Symbol;Acc:HGNC:5048] | 0.21 | 6.80 | 0.03 | -5.03913656828233 | 1.09e-10 | 2.52e-08 | down | yes |
| 37113 | NA | chr12 | + | 3077355 | 3286564 | ENST00000537971 | 8 | 4284 | ENSG00000011105 | TSPAN9 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005886(plasma membrane);GO:0005925(focal adhesion);GO:0005887(integral component of plasma membrane);GO:0003674(molecular_function);GO:0008150(biological_process);GO:0007166(cell surface receptor signaling pathway);GO:0097197(tetraspanin-enriched microdomain) | NA | NA | NA | tetraspanin 9 [Source:HGNC Symbol;Acc:HGNC:21640] | 14.396567 | 0.47 | 30.6531682493719 | 4.94 | 1.42e-10 | 3.25e-08 | up | yes |
| 35185 | NA | chr11 | - | 117820070 | 117828698 | ENST00000528014 | 7 | 672 | ENSG00000137731 | FXYD2 | GO:0006810(transport);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005216(ion channel activity);GO:0006811(ion transport);GO:0006813(potassium ion transport);GO:0006814(sodium ion transport);GO:0005886(plasma membrane);GO:0010248(establishment or maintenance of transmembrane electrochemical gradient);GO:0005887(integral component of plasma membrane);GO:0017080(sodium channel regulator activity);GO:2000649(regulation of sodium ion transmembrane transporter activity);GO:0070062(extracellular exosome);GO:0034220(ion transmembrane transport);GO:1903779(regulation of cardiac conduction);GO:0005890(sodium:potassium-exchanging ATPase complex);GO:0005215(transporter activity);GO:0005391(sodium:potassium-exchanging ATPase activity);GO:0036376(sodium ion export from cell);GO:1990573(potassium ion import across plasma membrane);GO:0090662(ATP hydrolysis coupled transmembrane transport);GO:0001558(regulation of cell growth);GO:0042127(regulation of cell proliferation);GO:0016323(basolateral plasma membrane);GO:0043231(intracellular membrane-bounded organelle) | 04024(cAMP signaling pathway);04022(cGMP - PKG signaling pathway);04911(Insulin secretion);04918(Thyroid hormone synthesis);04919(Thyroid hormone signaling pathway);04260(Cardiac muscle contraction);04261(Adrenergic signaling in cardiomyocytes);04970(Salivary secretion);04972(Pancreatic secretion);04976(Bile secretion);04973(Carbohydrate digestion and absorption);04974(Protein digestion and absorption);04978(Mineral absorption);04960(Aldosterone-regulated sodium reabsorption);04961(Endocrine and other factor-regulated calcium reabsorption);04964(Proximal tubule bicarbonate reclamation) | NA | NA | FXYD domain containing ion transport regulator 2 [Source:HGNC Symbol;Acc:HGNC:4026] | 1.99 | 64.206131 | 0.03 | -5.01395943677157 | 1.54e-10 | 3.47e-08 | down | yes |
| 126991 | NA | chr22 | - | 17592136 | 17628818 | ENST00000253413 | 9 | 1402 | ENSG00000131100 | ATP6V1E1 | GO:0006810(transport);GO:0016787(hydrolase activity);GO:0005765(lysosomal membrane);GO:0005829(cytosol);GO:0006811(ion transport);GO:0015991(ATP hydrolysis coupled proton transport);GO:0015992(proton transport);GO:0046961(proton-transporting ATPase activity, rotational mechanism);GO:0005515(protein binding);GO:0033178(proton-transporting two-sector ATPase complex, catalytic domain);GO:0005768(endosome);GO:0070062(extracellular exosome);GO:0034220(ion transmembrane transport);GO:0016324(apical plasma membrane);GO:0008286(insulin receptor signaling pathway);GO:0016241(regulation of macroautophagy);GO:0033572(transferrin transport);GO:0090383(phagosome acidification);GO:0051117(ATPase binding);GO:0016469(proton-transporting two-sector ATPase complex);GO:0008553(hydrogen-exporting ATPase activity, phosphorylative mechanism);GO:0005737(cytoplasm);GO:0005739(mitochondrion);GO:0005902(microvillus) | 00190(Oxidative phosphorylation);04150(mTOR signaling pathway);04145(Phagosome);04966(Collecting duct acid secretion);04721(Synaptic vesicle cycle);05323(Rheumatoid arthritis);05110(Vibrio cholerae infection);05120(Epithelial cell signaling in Helicobacter pylori infection) | K02150 | NA | ATPase H+ transporting V1 subunit E1 [Source:HGNC Symbol;Acc:HGNC:857] | 13.826056 | 0.43 | 32.339835892254 | 5.02 | 1.55e-10 | 3.47e-08 | up | yes |
| 108372 | NA | chr2 | + | 47402969 | 47483228 | ENST00000233146 | 16 | 3307 | ENSG00000095002 | MSH2 | GO:0005634(nucleus);GO:0016020(membrane);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0003677(DNA binding);GO:0005515(protein binding);GO:0006281(DNA repair);GO:0006974(cellular response to DNA damage stimulus);GO:0030983(mismatched DNA binding);GO:0006298(mismatch repair);GO:0032301(MutSalpha complex);GO:0005654(nucleoplasm);GO:0019901(protein kinase binding);GO:0006302(double-strand break repair);GO:0031573(intra-S DNA damage checkpoint);GO:0000784(nuclear chromosome, telomeric region);GO:0008022(protein C-terminus binding);GO:0032300(mismatch repair complex);GO:0019899(enzyme binding);GO:0010165(response to X-ray);GO:0008094(DNA-dependent ATPase activity);GO:0043524(negative regulation of neuron apoptotic process);GO:0030183(B cell differentiation);GO:0042803(protein homodimerization activity);GO:0003697(single-stranded DNA binding);GO:0032405(MutLalpha complex binding);GO:0016887(ATPase activity);GO:0008584(male gonad development);GO:0032139(dinucleotide insertion or deletion binding);GO:0032142(single guanine insertion binding);GO:0032181(dinucleotide repeat insertion binding);GO:0000403(Y-form DNA binding);GO:0000404(heteroduplex DNA loop binding);GO:0000406(double-strand/single-strand DNA junction binding);GO:0043570(maintenance of DNA repeat elements);GO:0045910(negative regulation of DNA recombination);GO:0051096(positive regulation of helicase activity);GO:0032302(MutSbeta complex);GO:0000287(magnesium ion binding);GO:0016446(somatic hypermutation of immunoglobulin genes);GO:0000400(four-way junction DNA binding);GO:0003690(double-stranded DNA binding);GO:0032137(guanine/thymine mispair binding);GO:0032143(single thymine insertion binding);GO:0032357(oxidized purine DNA binding);GO:0043531(ADP binding);GO:0003684(damaged DNA binding);GO:0019237(centromeric DNA binding);GO:0001701(in utero embryonic development);GO:0002204(somatic recombination of immunoglobulin genes involved in immune response);GO:0006119(oxidative phosphorylation);GO:0006301(postreplication repair);GO:0006311(meiotic gene conversion);GO:0007050(cell cycle arrest);GO:0007281(germ cell development);GO:0008340(determination of adult lifespan);GO:0008630(intrinsic apoptotic signaling pathway in response to DNA damage);GO:0010224(response to UV-B);GO:0016447(somatic recombination of immunoglobulin gene segments);GO:0019724(B cell mediated immunity);GO:0042771(intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator);GO:0045128(negative regulation of reciprocal meiotic recombination);GO:0045190(isotype switching);GO:0048298(positive regulation of isotype switching to IgA isotypes);GO:0048304(positive regulation of isotype switching to IgG isotypes) | 03430(Mismatch repair);05200(Pathways in cancer);05210(Colorectal cancer);01524(Platinum drug resistance) | K08735 | NA | mutS homolog 2 [Source:HGNC Symbol;Acc:HGNC:7325] | 10.005642 | 0.32 | 30.8079193287661 | 4.95 | 1.69e-10 | 3.72e-08 | up | yes |
| 194805 | NA | chrX | + | 47222853 | 47228990 | ENST00000428400 | 11 | 3016 | ENSG00000102225 | CDK16 | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0000166(nucleotide binding);GO:0004672(protein kinase activity);GO:0004674(protein serine/threonine kinase activity);GO:0005524(ATP binding);GO:0006468(protein phosphorylation);GO:0005829(cytosol);GO:0016740(transferase activity);GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0005886(plasma membrane);GO:0015630(microtubule cytoskeleton);GO:0005515(protein binding);GO:0030054(cell junction);GO:0045202(synapse);GO:0030154(cell differentiation);GO:0031410(cytoplasmic vesicle);GO:0007283(spermatogenesis);GO:0006887(exocytosis);GO:0031234(extrinsic component of cytoplasmic side of plasma membrane);GO:0043005(neuron projection);GO:0031175(neuron projection development);GO:0008021(synaptic vesicle);GO:0030133(transport vesicle);GO:0061178(regulation of insulin secretion involved in cellular response to glucose stimulus);GO:0030252(growth hormone secretion) | NA | NA | NA | cyclin dependent kinase 16 [Source:HGNC Symbol;Acc:HGNC:8749] | 12.611687 | 0.41 | 30.5179296076273 | 4.93 | 1.83e-10 | 3.99e-08 | up | yes |
| 170601 | NA | chr7 | - | 22934211 | 23014129 | ENST00000432176 | 11 | 13177 | ENSG00000122591 | FAM126A | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0005829(cytosol);GO:0005886(plasma membrane);GO:0004871(signal transducer activity);GO:0007165(signal transduction);GO:0005515(protein binding);GO:0090002(establishment of protein localization to plasma membrane);GO:0042552(myelination);GO:0046854(phosphatidylinositol phosphorylation);GO:0043005(neuron projection) | NA | NA | NA | family with sequence similarity 126 member A [Source:HGNC Symbol;Acc:HGNC:24587] | 2.13 | 0.07 | 30.8708228270006 | 4.95 | 1.87e-10 | 4.02e-08 | up | yes |
| 79055 | NA | chr17 | + | 16439063 | 16441861 | ENST00000581913 | 4 | 1091 | ENSG00000175061 | LRRC75A-AS1 | NA | NA | NA | NA | LRRC75A antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:28619] | 1.08 | 33.861019 | 0.03 | -4.97517568167784 | 1.99e-10 | 4.24e-08 | down | yes |
| 192124 | NA | chr9 | + | 129119022 | 129131639 | ENST00000417504 | 5 | 567 | ENSG00000119383 | PTPA | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0016853(isomerase activity);GO:0005654(nucleoplasm);GO:0003755(peptidyl-prolyl cis-trans isomerase activity);GO:0005515(protein binding);GO:0000413(protein peptidyl-prolyl isomerization);GO:0070062(extracellular exosome);GO:0043065(positive regulation of apoptotic process);GO:0019211(phosphatase activator activity);GO:0005102(receptor binding);GO:0034704(calcium channel complex);GO:0042803(protein homodimerization activity);GO:0016887(ATPase activity);GO:0046982(protein heterodimerization activity);GO:0008160(protein tyrosine phosphatase activator activity);GO:0019888(protein phosphatase regulator activity);GO:0051721(protein phosphatase 2A binding);GO:0030472(mitotic spindle organization in nucleus);GO:0032515(negative regulation of phosphoprotein phosphatase activity);GO:0032516(positive regulation of phosphoprotein phosphatase activity);GO:0035307(positive regulation of protein dephosphorylation);GO:0035308(negative regulation of protein dephosphorylation);GO:0043666(regulation of phosphoprotein phosphatase activity);GO:0000159(protein phosphatase type 2A complex);GO:0043085(positive regulation of catalytic activity) | 04931(Insulin resistance) | NA | NA | protein phosphatase 2 phosphatase activator [Source:HGNC Symbol;Acc:HGNC:9308] | 33.036552 | 1.06 | 31.1739402443409 | 4.96 | 2.15e-10 | 4.52e-08 | up | yes |
| 115532 | NA | chr2 | - | 177230305 | 177265131 | ENST00000397062 | 5 | 2853 | ENSG00000116044 | NFE2L2 | GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0005634(nucleus);GO:0071356(cellular response to tumor necrosis factor);GO:2000352(negative regulation of endothelial cell apoptotic process);GO:0005737(cytoplasm);GO:0005794(Golgi apparatus);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0006355(regulation of transcription, DNA-templated);GO:0006351(transcription, DNA-templated);GO:0016567(protein ubiquitination);GO:0003677(DNA binding);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0005886(plasma membrane);GO:0005813(centrosome);GO:0005515(protein binding);GO:0034599(cellular response to oxidative stress);GO:0019904(protein domain specific binding);GO:0006366(transcription from RNA polymerase II promoter);GO:0043161(proteasome-mediated ubiquitin-dependent protein catabolic process);GO:0001102(RNA polymerase II activating transcription factor binding);GO:0010499(proteasomal ubiquitin-independent protein catabolic process);GO:1903206(negative regulation of hydrogen peroxide-induced cell death);GO:0071498(cellular response to fluid shear stress);GO:0071499(cellular response to laminar fluid shear stress);GO:1903071(positive regulation of ER-associated ubiquitin-dependent protein catabolic process);GO:0000976(transcription regulatory region sequence-specific DNA binding);GO:0010628(positive regulation of gene expression);GO:1902176(negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway);GO:0036499(PERK-mediated unfolded protein response);GO:0070301(cellular response to hydrogen peroxide);GO:0044212(transcription regulatory region DNA binding);GO:0036003(positive regulation of transcription from RNA polymerase II promoter in response to stress);GO:0032993(protein-DNA complex);GO:0030968(endoplasmic reticulum unfolded protein response);GO:0000980(RNA polymerase II distal enhancer sequence-specific DNA binding);GO:0001205(transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding);GO:0001221(transcription cofactor binding);GO:0008134(transcription factor binding);GO:0043565(sequence-specific DNA binding);GO:0006954(inflammatory response);GO:0007568(aging);GO:0010667(negative regulation of cardiac muscle cell apoptotic process);GO:0010976(positive regulation of neuron projection development);GO:0030194(positive regulation of blood coagulation);GO:0034976(response to endoplasmic reticulum stress);GO:0035690(cellular response to drug);GO:0036091(positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress);GO:0042149(cellular response to glucose starvation);GO:0043536(positive regulation of blood vessel endothelial cell migration);GO:0045454(cell redox homeostasis);GO:0045766(positive regulation of angiogenesis);GO:0045893(positive regulation of transcription, DNA-templated);GO:0045995(regulation of embryonic development);GO:0046223(aflatoxin catabolic process);GO:0046326(positive regulation of glucose import);GO:0060548(negative regulation of cell death);GO:0071456(cellular response to hypoxia);GO:1902037(negative regulation of hematopoietic stem cell differentiation);GO:1903788(positive regulation of glutathione biosynthetic process);GO:1904753(negative regulation of vascular associated smooth muscle cell migration);GO:2000121(regulation of removal of superoxide radicals);GO:2000379(positive regulation of reactive oxygen species metabolic process);GO:0000785(chromatin) | 04141(Protein processing in endoplasmic reticulum);05200(Pathways in cancer);05225(Hepatocellular carcinoma);05418(Fluid shear stress and atherosclerosis) | K05638 | NA | nuclear factor, erythroid 2 like 2 [Source:HGNC Symbol;Acc:HGNC:7782] | 25.017759 | 0.86 | 29.2198682535419 | 4.87 | 2.18e-10 | 4.52e-08 | up | yes |
| 9947 | NA | chr1 | - | 145854406 | 145859779 | ENST00000369299 | 6 | 1235 | ENSG00000131788 | PIAS3 | GO:0008270(zinc ion binding);GO:0019789(SUMO transferase activity);GO:0016925(protein sumoylation);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016607(nuclear speck);GO:0046872(metal ion binding);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0016874(ligase activity);GO:0005515(protein binding);GO:0065009(regulation of molecular function);GO:0005654(nucleoplasm);GO:0008022(protein C-terminus binding);GO:0019899(enzyme binding);GO:0033235(positive regulation of protein sumoylation);GO:0033234(negative regulation of protein sumoylation);GO:0015459(potassium channel regulator activity);GO:0047485(protein N-terminus binding);GO:0009725(response to hormone);GO:0010628(positive regulation of gene expression);GO:0045838(positive regulation of membrane potential);GO:0030425(dendrite);GO:0045202(synapse) | 04120(Ubiquitin mediated proteolysis);04630(Jak-STAT signaling pathway) | NA | NA | protein inhibitor of activated STAT 3 [Source:HGNC Symbol;Acc:HGNC:16861] | 16.40592 | 0.53 | 30.6920046245889 | 4.94 | 2.24e-10 | 4.61e-08 | up | yes |
| 95711 | NA | chr19 | - | 11342776 | 11346305 | ENST00000447337 | 4 | 916 | ENSG00000105518 | TMEM205 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0070062(extracellular exosome) | NA | NA | NA | transmembrane protein 205 [Source:HGNC Symbol;Acc:HGNC:29631] | 0.78 | 24.702267 | 0.03 | -4.97976557824233 | 2.57e-10 | 5.20e-08 | down | yes |
| 108389 | NA | chr2 | + | 47783082 | 47806945 | ENST00000540021 | 8 | 3930 | ENSG00000116062 | MSH6 | GO:0005634(nucleus);GO:0005794(Golgi apparatus);GO:0043231(intracellular membrane-bounded organelle);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0003677(DNA binding);GO:0005694(chromosome);GO:0005515(protein binding);GO:0006281(DNA repair);GO:0006974(cellular response to DNA damage stimulus);GO:0030983(mismatched DNA binding);GO:0006298(mismatch repair);GO:0032301(MutSalpha complex);GO:0016032(viral process);GO:0035064(methylated histone binding);GO:0008094(DNA-dependent ATPase activity);GO:0008340(determination of adult lifespan);GO:0008630(intrinsic apoptotic signaling pathway in response to DNA damage);GO:0032405(MutLalpha complex binding);GO:0016887(ATPase activity);GO:0032142(single guanine insertion binding);GO:0045910(negative regulation of DNA recombination);GO:0051096(positive regulation of helicase activity);GO:0009411(response to UV);GO:0000287(magnesium ion binding);GO:0016446(somatic hypermutation of immunoglobulin genes);GO:0000400(four-way junction DNA binding);GO:0003690(double-stranded DNA binding);GO:0032137(guanine/thymine mispair binding);GO:0032143(single thymine insertion binding);GO:0032357(oxidized purine DNA binding);GO:0043531(ADP binding);GO:0016447(somatic recombination of immunoglobulin gene segments);GO:0045190(isotype switching);GO:0000710(meiotic mismatch repair);GO:0097193(intrinsic apoptotic signaling pathway);GO:0003682(chromatin binding);GO:0003684(damaged DNA binding);GO:0045830(positive regulation of isotype switching);GO:0000790(nuclear chromatin) | 03430(Mismatch repair);05200(Pathways in cancer);05210(Colorectal cancer);01524(Platinum drug resistance) | K08737 | NA | mutS homolog 6 [Source:HGNC Symbol;Acc:HGNC:7329] | 23.162136 | 0.81 | 28.5278194488579 | 4.83 | 2.59e-10 | 5.20e-08 | up | yes |
| 20758 | NA | chr10 | - | 70298973 | 70381914 | ENST00000355790 | 5 | 3414 | ENSG00000172731 | LRRC20 | GO:0005515(protein binding) | NA | NA | NA | leucine rich repeat containing 20 [Source:HGNC Symbol;Acc:HGNC:23421] | 1.28 | 37.883965 | 0.03 | -4.88413482367765 | 2.80e-10 | 5.57e-08 | down | yes |
| 192868 | NA | chr9 | - | 136440097 | 136477689 | ENST00000313050 | 30 | 8806 | ENSG00000148396 | SEC16A | GO:0048208(COPII vesicle coating);GO:0000139(Golgi membrane);GO:0006810(transport);GO:0016020(membrane);GO:0015031(protein transport);GO:0005794(Golgi apparatus);GO:0005783(endoplasmic reticulum);GO:0005829(cytosol);GO:0005789(endoplasmic reticulum membrane);GO:0005515(protein binding);GO:0016192(vesicle-mediated transport);GO:0021762(substantia nigra development);GO:0007029(endoplasmic reticulum organization) | NA | NA | NA | SEC16 homolog A, endoplasmic reticulum export factor [Source:HGNC Symbol;Acc:HGNC:29006] | 0.95 | 27.602127 | 0.03 | -4.86212265503282 | 2.97e-10 | 5.83e-08 | down | yes |
| 171991 | NA | chr7 | + | 44606591 | 44708328 | ENST00000449767 | 23 | 3479 | ENSG00000105953 | OGDH | GO:0055114(oxidation-reduction process);GO:0006099(tricarboxylic acid cycle);GO:0005739(mitochondrion);GO:0008152(metabolic process);GO:0016624(oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor);GO:0004591(oxoglutarate dehydrogenase (succinyl-transferring) activity);GO:0030976(thiamine pyrophosphate binding);GO:0046872(metal ion binding);GO:0016491(oxidoreductase activity);GO:0006096(glycolytic process);GO:0005759(mitochondrial matrix);GO:0031966(mitochondrial membrane);GO:0006554(lysine catabolic process);GO:0034641(cellular nitrogen compound metabolic process);GO:0006091(generation of precursor metabolites and energy);GO:0045252(oxoglutarate dehydrogenase complex);GO:0031072(heat shock protein binding);GO:0034602(oxoglutarate dehydrogenase (NAD+) activity);GO:0051087(chaperone binding);GO:0006103(2-oxoglutarate metabolic process);GO:0006104(succinyl-CoA metabolic process);GO:0006734(NADH metabolic process);GO:0021695(cerebellar cortex development);GO:0021756(striatum development);GO:0021766(hippocampus development);GO:0021794(thalamus development);GO:0021860(pyramidal neuron development);GO:0022028(tangential migration from the subventricular zone to the olfactory bulb);GO:0061034(olfactory bulb mitral cell layer development) | 00020(Citrate cycle (TCA cycle));00310(Lysine degradation);00380(Tryptophan metabolism) | K00164 | EC:1.2.4.2 | oxoglutarate dehydrogenase [Source:HGNC Symbol;Acc:HGNC:8124] | 8.96 | 0.31 | 28.4908929110585 | 4.83 | 3.46e-10 | 6.72e-08 | up | yes |
| 68330 | NA | chr16 | - | 4463568 | 4469365 | ENST00000576176 | 3 | 672 | ENSG00000153406 | NMRAL1 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0048471(perinuclear region of cytoplasm);GO:0005829(cytosol);GO:0000050(urea cycle) | NA | NA | NA | NmrA like redox sensor 1 [Source:HGNC Symbol;Acc:HGNC:24987] | 1.36 | 41.062065 | 0.03 | -4.91556868937956 | 3.62e-10 | 6.96e-08 | down | yes |
| 94968 | NA | chr19 | + | 8418405 | 8439017 | ENST00000602117 | 5 | 1667 | ENSG00000099785 | MARCH2 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0008270(zinc ion binding);GO:0046872(metal ion binding);GO:0016740(transferase activity);GO:0016567(protein ubiquitination);GO:0005765(lysosomal membrane);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0005515(protein binding);GO:0031410(cytoplasmic vesicle);GO:0005764(lysosome);GO:0005768(endosome);GO:0006897(endocytosis);GO:0010008(endosome membrane);GO:0004842(ubiquitin-protein transferase activity) | NA | NA | NA | membrane associated ring-CH-type finger 2 [Source:HGNC Symbol;Acc:HGNC:28038] | 0.39 | 11.77881 | 0.03 | -4.93416515262289 | 3.78e-10 | 7.20e-08 | down | yes |
| 190398 | NA | chr9 | - | 100302084 | 100352939 | ENST00000374902 | 15 | 3074 | ENSG00000136891 | TEX10 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005515(protein binding);GO:0005654(nucleoplasm);GO:0005730(nucleolus);GO:0031965(nuclear membrane);GO:0071339(MLL1 complex);GO:0006364(rRNA processing);GO:0097344(Rix1 complex) | NA | NA | NA | testis expressed 10 [Source:HGNC Symbol;Acc:HGNC:25988] | 0.56 | 16.043062 | 0.03 | -4.83909906134783 | 4.43e-10 | 8.35e-08 | down | yes |
| 120342 | NA | chr20 | + | 2655413 | 2658392 | ENST00000612233 | 1 | 2980 | ENSG00000101361 | NOP56 | GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0030529(intracellular ribonucleoprotein complex);GO:0001650(fibrillar center);GO:0005654(nucleoplasm);GO:0042254(ribosome biogenesis);GO:0005730(nucleolus);GO:0032040(small-subunit processome);GO:0005515(protein binding);GO:0045296(cadherin binding);GO:0031428(box C/D snoRNP complex);GO:0006364(rRNA processing);GO:0030515(snoRNA binding);GO:1990226(histone methyltransferase binding);GO:0000154(rRNA modification);GO:0005732(small nucleolar ribonucleoprotein complex);GO:0070761(pre-snoRNP complex) | 03008(Ribosome biogenesis in eukaryotes) | NA | NA | NOP56 ribonucleoprotein [Source:HGNC Symbol;Acc:HGNC:15911] | 0.19 | 5.73 | 0.03 | -4.90975496656206 | 4.81e-10 | 8.96e-08 | down | yes |
| 191802 | NA | chr9 | - | 128166065 | 128191568 | ENST00000372954 | 17 | 2731 | ENSG00000148337 | CIZ1 | GO:0005634(nucleus);GO:0003676(nucleic acid binding);GO:0008270(zinc ion binding);GO:0046872(metal ion binding);GO:0005654(nucleoplasm);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0030332(cyclin binding);GO:0032298(positive regulation of DNA-dependent DNA replication initiation);GO:0051457(maintenance of protein location in nucleus) | NA | NA | NA | CDKN1A interacting zinc finger protein 1 [Source:HGNC Symbol;Acc:HGNC:16744] | 1.23 | 34.206684 | 0.04 | -4.80265811579905 | 4.91e-10 | 9.07e-08 | down | yes |
Definition of table:
| term | 意义 |
|---|---|
| chr | The name of the chromosome (e.g. chr3, chrY, chr2 random) or scaffold (e.g. scaffold10671) |
| strand | sense strand="+"; antisense strand="-" |
| start | The starting position of transcript in the chromosome or scaffold |
| end | The ending position of transcript in the chromosome or scaffold |
| t_name | The name of transcript |
| num_exons | number of exons |
| length | length of transcript |
| gene_id | The ID of gene, some ids started with "MSTRG" are named by transcript assembly software StringTie |
| gene_name | The name of gene (gene symbol) |
| GO | Gene ontology term and description http://www.geneontology.org/ |
| KEGG | KEGG pathway annotation and description http://www.kegg.jp/kegg/kegg3a.html |
| KO_Entry | KO functional ortholog, KO groups identified by K numbers and, in metabolic maps |
| EC | enzyme |
| Description | Description of gene |
| cov.A | count number or coverage of transcript |
| FPKM.A | Normalization of gene expression level by FPKM value |
| fc | fold change |
| log2(fc) | log2 of fold change value |
| pval | p value |
| qval | q value or FDR value |
| regulation | up-regulation or down-regulation |
| significant | yes or no |
document location: summary/7_differential_expression/*VS*/*_Transcript_differential_expression.xlsx
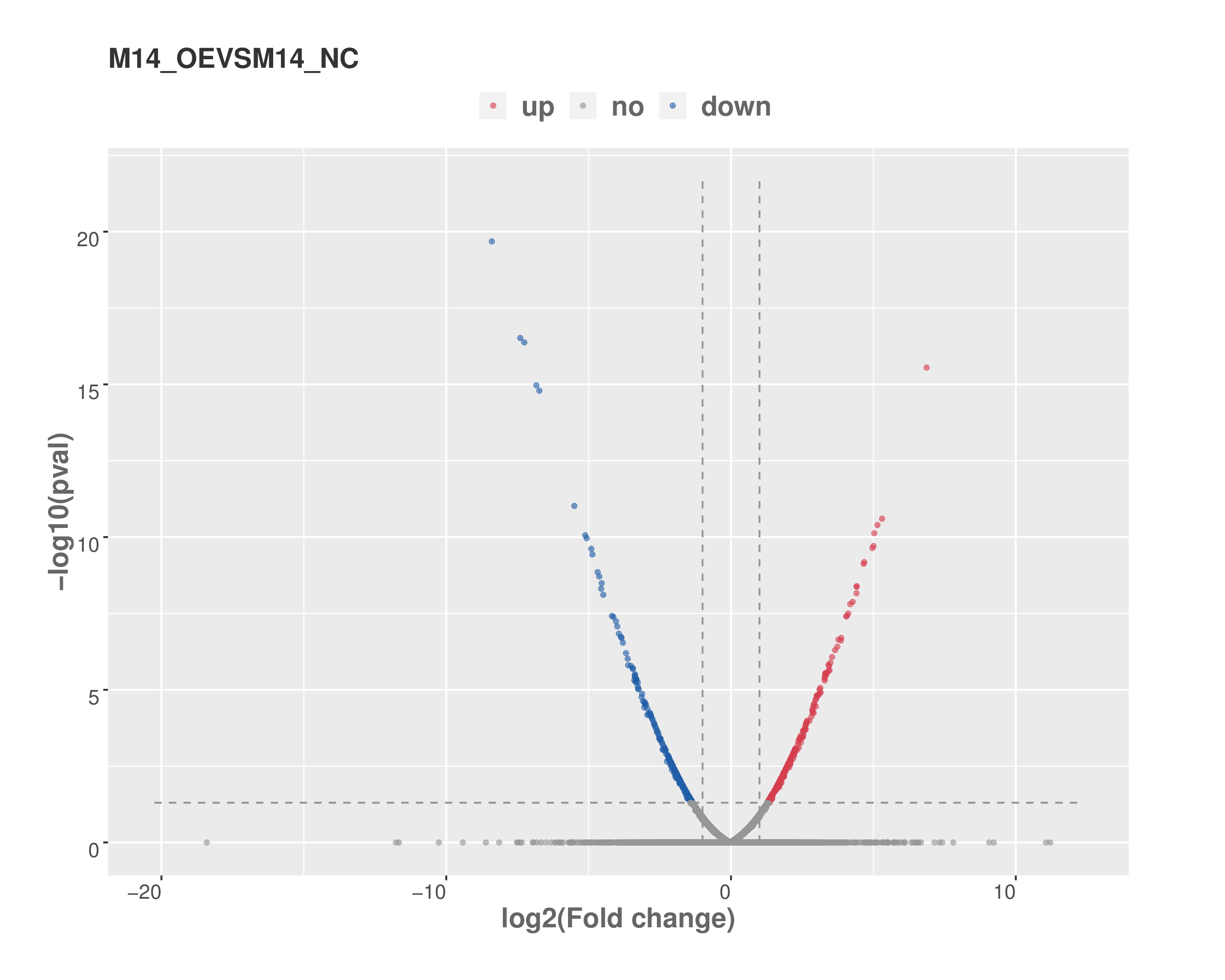
document location:
summary/7_differential_expression/*VS*/*_Genes_volcano.png
summary/7_differential_expression/*VS*/*_Transcripts_volcano.png
Heatmap is a graphical representation of data where the individual z values (repeated) or Log10 (non-repeated) contained in a matrix are represented as color scheme. Calculation formula of z value is shown below. Sample-i means expression level (fpkm value).
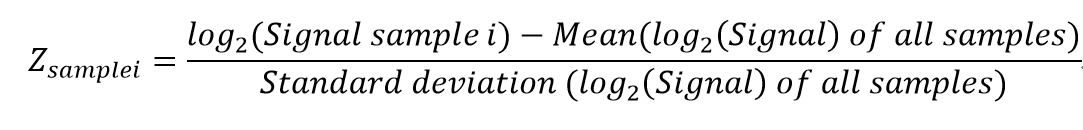
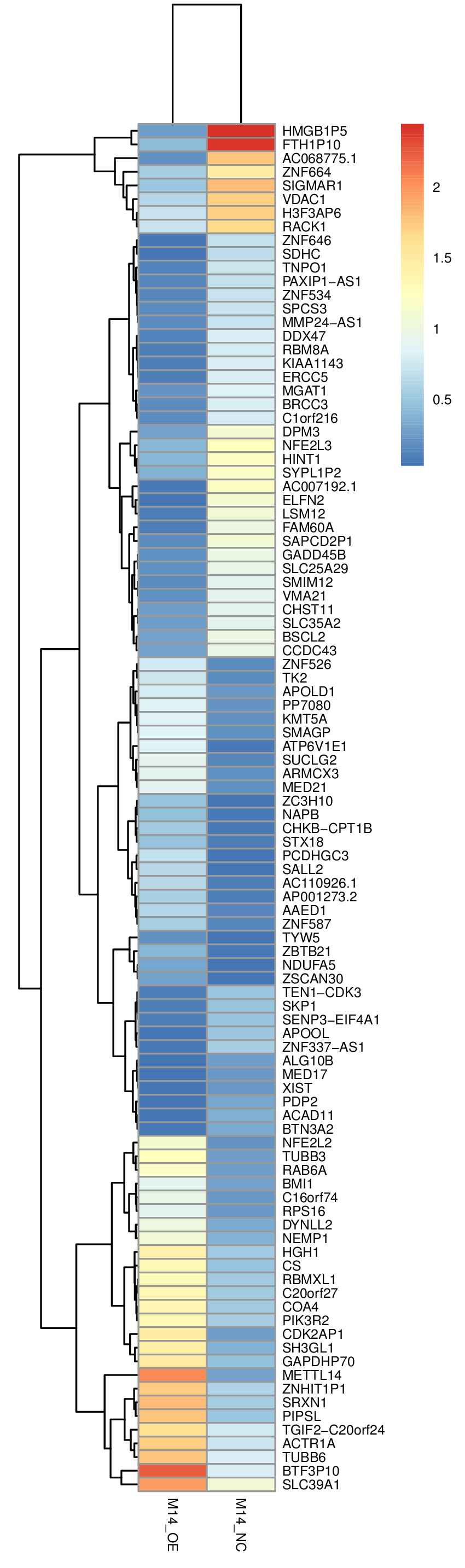
document location:
summary/7_differential_expression/*VS*/*_genes_heatmap.png
summary/7_differential_expression/*VS*/*_transcripts_heatmap.png
Gene ontology (GO) (http://www.geneontology.org) is a major bioinformatics initiative to unify the representation of gene and gene product attributes across all species. More specifically, the project aims to: 1) maintain and develop its controlled vocabulary of gene and gene product attributes; 2) annotate genes and gene products, and assimilate and disseminate annotation data; and 3) provide tools for easy access to all aspects of the data provided by the project, and to enable functional interpretation of experimental data using the GO, for example via enrichment analysis.
GO is part of a larger classification effort, the Open Biomedical Ontologies (OBO).
Although gene nomenclature itself aims to maintain and develop controlled vocabulary of gene and gene products, the Gene Ontology extends the effort by using markup language to make the data (not only of the genes and their products but also of all their attributes) machine readable, and to do so in a way that is unified across all species (whereas gene nomenclature conventions vary by biologic taxon).
Significant GO terms were calculated by Hypergeometric equation as shown below. TB gene number=number of total genes; TS gene number=number of differentially expressed genes in total genes; B gene number=total number of gene in all GO terms; S gene number=number of differentially expressed genes in this GO term. Those GO terms with p value<0.05 were defined as significant GO terms.
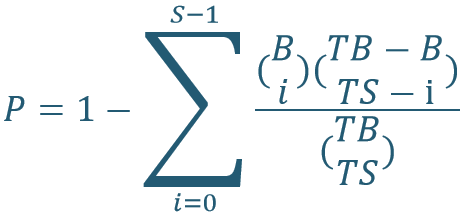
Significant GO terms:
| GO_ID | GO_Term | GO_function | GO_class | Genes | S gene number | TS gene number | B gene number | TB gene number | pvalue |
| GO:0036518 | chemorepulsion of dopaminergic neuron axon | NA | NA | RYK;WNT5A | 2 | 368 | 2 | 15874 | 0.00 |
| GO:0003697 | single-stranded DNA binding | molecular_function | 8 | BIVM-ERCC5;CNBP;ERCC5;POLR2D;RPA2;SSBP4;TWNK;XRCC2 | 8 | 368 | 86 | 15874 | 0.00 |
| GO:0016591 | DNA-directed RNA polymerase II, holoenzyme | cellular_component | 10 | ERCC5;GCOM1;RPRD1A | 3 | 368 | 10 | 15874 | 0.00 |
| GO:0031369 | translation initiation factor binding | molecular_function | 5 | CELF1;LARP1;POLR2D;ZPR1 | 4 | 368 | 22 | 15874 | 0.00 |
| GO:0030292 | protein tyrosine kinase inhibitor activity | molecular_function | 7 | FGFR1OP;RACK1 | 2 | 368 | 3 | 15874 | 0.00 |
| GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle | biological_process | 14 | ID2;ZPR1 | 2 | 368 | 3 | 15874 | 0.00 |
| GO:0034067 | protein localization to Golgi apparatus | biological_process | 8 | CSNK1D;PAQR3;RAB6A | 3 | 368 | 11 | 15874 | 0.00 |
| GO:0005634 | nucleus | cellular_component | 9 | ACAD11;AGO1;AHCY;AHRR;AKT3;ANAPC11;ANKRA2;AP001273.2;APC;API5;ARHGEF5;BAG4;BCL2L13;BIVM-ERCC5;BMI1;BOLA2B;BRCC3;C19orf68;C1orf109;CCNL1;CD3EAP;CDK11A;CDK15;CDK19;CDK2AP1;CELF1;CFL2;CITED2;CNBP;CREB3;CREB5;CSNK1D;CS;DCXR;DDX39A;DDX47;DNAJB12;DPH1;DYNLL2;ELOF1;EMG1;ERCC5;FAM58A;FAM60A;FGFR1OP;FOXD1;GADD45B;GDAP1;GLO1;GMEB2;H3F3A;HDGF;HIC2;HINT1;HJURP;HKR1;ID2;IDH3B;IER3;IMPDH1;INTS6;KLF6;KMT5A;LBH;LSM2;LSM5;MAPK7;MATR3;MCL1;MED17;MED21;MEF2B;MEMO1;METTL14;MRPL19;NEMP1;NFE2L2;NFE2L3;NFIC;NSRP1;NXN;PAWR;PCGF1;PCNP;PHLPP2;PIH1D1;PIK3R2;PNN;POLR2D;PPIA;PRKACA;PSMA1;PSMC5;RACK1;RBAK;RBBP4;RBM22;RBM3;RBM8A;RBMXL1;RERG;RNF126;RPA2;RPL10;RPL17;RYK;S100A16;SALL2;SAP30BP;SAP30;SCAND1;SDR39U1;SENP3-EIF4A1;SIGMAR1;SKP1;SLC35A2;SOX6;SP1;SRSF3;SSBP4;TAF11;TFDP1;TGIF2-C20orf24;TMEM250;TNIP2;TNPO1;TRIM11;TUBB3;TUBB6;TUB;UBE2V2;VDAC1;WDFY1;XRCC2;YAF2;YY1;ZBTB21;ZBTB8A;ZFP1;ZNF132;ZNF24;ZNF267;ZNF268;ZNF324;ZNF383;ZNF500;ZNF526;ZNF534;ZNF587;ZNF646;ZNF664;ZNF665;ZNF721;ZNF746;ZNF766;ZPR1;ZSCAN30;ZSCAN32 | 158 | 368 | 5658 | 15874 | 0.00 |
| GO:0005789 | endoplasmic reticulum membrane | cellular_component | 11 | ALG10B;BSCL2;CLN6;CLN8;COPZ1;CREB3;CYB5RL;DNAJB12;DPM3;EBPL;EXTL2;GJB1;GJC1;HACD2;ILDR2;KRTCAP2;KSR1;LCLAT1;MGST3;MPDU1;ORMDL3;PCYT1A;PGAP1;RAB6A;SEC61G;SIGMAR1;SLC39A1;SPCS3;STX18;TMEM208;TRIQK;UBE2J2;VMA21 | 33 | 368 | 846 | 15874 | 0.00 |
| GO:0031519 | PcG protein complex | cellular_component | 11 | BMI1;PCGF1;SKP1;YY1 | 4 | 368 | 26 | 15874 | 0.00 |
| GO:0061631 | ubiquitin conjugating enzyme activity | molecular_function | 7 | UBE2D2;UBE2D4;UBE2J2;UBE2V2 | 4 | 368 | 28 | 15874 | 0.00 |
| GO:0003676 | nucleic acid binding | molecular_function | 5 | AGO1;CELF1;CNBP;CREB5;DDX39A;DDX47;GPATCH4;HIC2;HKR1;IMPDH1;KLF6;MATR3;RBAK;RBM22;RBM3;RBM8A;RBMXL1;RBSN;SALL2;SP1;SRSF3;THAP6;YY1;ZBTB21;ZBTB8A;ZFP1;ZNF132;ZNF24;ZNF267;ZNF268;ZNF324;ZNF383;ZNF428;ZNF500;ZNF526;ZNF534;ZNF587;ZNF646;ZNF664;ZNF665;ZNF721;ZNF746;ZNF766;ZSCAN30;ZSCAN32 | 45 | 368 | 1311 | 15874 | 0.01 |
| GO:1990726 | Lsm1-7-Pat1 complex | NA | NA | LSM2;LSM5 | 2 | 368 | 5 | 15874 | 0.01 |
| GO:1904938 | planar cell polarity pathway involved in axon guidance | NA | NA | RYK;WNT5A | 2 | 368 | 5 | 15874 | 0.01 |
| GO:0006099 | tricarboxylic acid cycle | biological_process | 10 | CS;IDH3B;SDHC;SUCLG2 | 4 | 368 | 31 | 15874 | 0.01 |
| GO:0006895 | Golgi to endosome transport | biological_process | 7 | CORO7;RBSN;SYS1 | 3 | 368 | 17 | 15874 | 0.01 |
| GO:2000052 | positive regulation of non-canonical Wnt signaling pathway | biological_process | 11 | CSNK1D;WNT5A | 2 | 368 | 6 | 15874 | 0.01 |
| GO:0038202 | TORC1 signaling | biological_process | 9 | LARP1;MLST8 | 2 | 368 | 6 | 15874 | 0.01 |
| GO:0071806 | protein transmembrane transport | biological_process | 6 | MCL1;SEC61G | 2 | 368 | 6 | 15874 | 0.01 |
| GO:0036003 | positive regulation of transcription from RNA polymerase II promoter in response to stress | biological_process | 14 | MAPK7;NFE2L2 | 2 | 368 | 6 | 15874 | 0.01 |
| GO:0048524 | positive regulation of viral process | biological_process | 6 | STOM;TMEM250 | 2 | 368 | 7 | 15874 | 0.01 |
| GO:0097428 | protein maturation by iron-sulfur cluster transfer | biological_process | 7 | BOLA2B;ISCA2 | 2 | 368 | 7 | 15874 | 0.01 |
| GO:0031931 | TORC1 complex | cellular_component | 8 | LARP1;MLST8 | 2 | 368 | 7 | 15874 | 0.01 |
| GO:0034115 | negative regulation of heterotypic cell-cell adhesion | biological_process | 7 | MAPK7;MYADM | 2 | 368 | 7 | 15874 | 0.01 |
| GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain | biological_process | 15 | CCNL1;FAM58A | 2 | 368 | 7 | 15874 | 0.01 |
| GO:0017183 | peptidyl-diphthamide biosynthetic process from peptidyl-histidine | biological_process | 6 | DPH1;DPH2 | 2 | 368 | 7 | 15874 | 0.01 |
| GO:0071499 | cellular response to laminar fluid shear stress | biological_process | 7 | MAPK7;NFE2L2 | 2 | 368 | 7 | 15874 | 0.01 |
| GO:1904948 | midbrain dopaminergic neuron differentiation | NA | NA | RYK;WNT5A | 2 | 368 | 7 | 15874 | 0.01 |
| GO:0016264 | gap junction assembly | biological_process | 8 | GJB1;GJC1 | 2 | 368 | 7 | 15874 | 0.01 |
| GO:1903358 | regulation of Golgi organization | biological_process | 6 | RBSN;STX18 | 2 | 368 | 7 | 15874 | 0.01 |
| GO:0007017 | microtubule-based process | biological_process | 5 | DYNLL2;TUBA4A;TUBB3;TUBB6 | 4 | 368 | 38 | 15874 | 0.01 |
| GO:0050804 | modulation of synaptic transmission | biological_process | 8 | HRH1;PRKACA;PSMC5 | 3 | 368 | 21 | 15874 | 0.01 |
| GO:0043066 | negative regulation of apoptotic process | biological_process | 9 | API5;BAG4;CHST11;CITED2;DDAH2;ERCC5;GLO1;IER3;LAMTOR5;MAPK7;MCL1;PLK2;RPL10;SEMA4D;SOCS2;TXNDC5;WNT5A;ZNF268 | 18 | 368 | 432 | 15874 | 0.01 |
| GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding | molecular_function | 11 | AGO1;CREB3;FOXD1;GMEB2;KLF6;MEF2B;NFE2L3;NFIC;RBBP4;SOX6;SP1;SSBP4;YY1;ZNF746 | 14 | 368 | 306 | 15874 | 0.01 |
| GO:0005688 | U6 snRNP | cellular_component | 9 | LSM2;LSM5 | 2 | 368 | 8 | 15874 | 0.01 |
| GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion | biological_process | 12 | BIVM-ERCC5;ERCC5;RPA2 | 3 | 368 | 22 | 15874 | 0.01 |
| GO:0004520 | endodeoxyribonuclease activity | molecular_function | 8 | BIVM-ERCC5;ERCC5;XRCC2 | 3 | 368 | 22 | 15874 | 0.01 |
| GO:0003735 | structural constituent of ribosome | molecular_function | 4 | MRPL19;MRPL35;MRPL49;MRPS33;NDUFA7;RPL10;RPL17;RPS16;RPS5 | 9 | 368 | 163 | 15874 | 0.01 |
| GO:0006289 | nucleotide-excision repair | biological_process | 10 | BIVM-ERCC5;ERCC5;ORAOV1;RPA2 | 4 | 368 | 41 | 15874 | 0.01 |
| GO:0030163 | protein catabolic process | biological_process | 7 | AC068580.4;CLN6;CLN8;PSMC5 | 4 | 368 | 41 | 15874 | 0.01 |
| GO:0006355 | regulation of transcription, DNA-templated | biological_process | 11 | AGO1;AHRR;BMI1;CCNL1;CDK11A;CITED2;CNBP;CREB3;CREB5;ELOF1;FOXD1;GMEB2;HDGF;HIC2;HINT1;HKR1;ID2;IGF2;KLF6;KMT5A;LBH;MED17;MED21;MEF2B;NFE2L2;NFE2L3;NFIC;PAWR;PCGF1;PIH1D1;PNN;RBAK;RBBP4;RNF141;SALL2;SAP30BP;SAP30;SCAND1;SOX6;SP1;TAF11;TFDP1;TGIF2-C20orf24;TNIP2;YAF2;YY1;ZBTB21;ZBTB8A;ZFP1;ZNF132;ZNF24;ZNF267;ZNF268;ZNF324;ZNF383;ZNF500;ZNF526;ZNF534;ZNF587;ZNF646;ZNF664;ZNF665;ZNF721;ZNF746;ZNF766;ZSCAN30;ZSCAN32 | 67 | 368 | 2232 | 15874 | 0.01 |
| GO:0030529 | intracellular ribonucleoprotein complex | cellular_component | 7 | AGO1;CELF1;LSM2;LSM5;MRPL19;MRPL35;MRPL49;MRPS33;NSRP1;RACK1;RBMXL1;RPL10;RPL17;RPS16;RPS5 | 15 | 368 | 345 | 15874 | 0.02 |
| GO:0098869 | cellular oxidant detoxification | NA | NA | AAED1;GPX2;MGST3;NXN;SRXN1 | 5 | 368 | 64 | 15874 | 0.02 |
| GO:0032886 | regulation of microtubule-based process | biological_process | 6 | APC;MEMO1 | 2 | 368 | 9 | 15874 | 0.02 |
| GO:0034498 | early endosome to Golgi transport | biological_process | 8 | RAB6A;RBSN | 2 | 368 | 9 | 15874 | 0.02 |
| GO:0048843 | negative regulation of axon extension involved in axon guidance | biological_process | 6 | RYK;WNT5A | 2 | 368 | 9 | 15874 | 0.02 |
| GO:0032486 | Rap protein signal transduction | biological_process | 10 | PLK2;SGSM3 | 2 | 368 | 9 | 15874 | 0.02 |
| GO:2000352 | negative regulation of endothelial cell apoptotic process | biological_process | 11 | MAPK7;NFE2L2;TNIP2 | 3 | 368 | 24 | 15874 | 0.02 |
| GO:0005921 | gap junction | cellular_component | 5 | GJB1;GJC1;SGSM3 | 3 | 368 | 24 | 15874 | 0.02 |
| GO:0006351 | transcription, DNA-templated | biological_process | 10 | AGO1;AHRR;BMI1;CCNL1;CITED2;CNBP;CREB3;CREB5;ELOF1;FOXD1;GMEB2;HDGF;HIC2;HINT1;HKR1;ID2;KLF6;KMT5A;LBH;MED17;MED21;MEF2B;NFE2L2;NFE2L3;NFIC;PAWR;PCGF1;PIH1D1;PNN;POLR2D;RBAK;RBBP4;SALL2;SAP30BP;SAP30;SOX6;SP1;TAF11;TFDP1;TNIP2;YAF2;YY1;ZBTB21;ZBTB8A;ZFP1;ZNF132;ZNF24;ZNF267;ZNF268;ZNF324;ZNF383;ZNF500;ZNF526;ZNF534;ZNF587;ZNF646;ZNF664;ZNF665;ZNF721;ZNF746;ZNF766;ZSCAN30;ZSCAN32 | 63 | 368 | 2105 | 15874 | 0.02 |
| GO:0017070 | U6 snRNA binding | molecular_function | 8 | LSM2;RBM22 | 2 | 368 | 10 | 15874 | 0.02 |
| GO:0010225 | response to UV-C | biological_process | 8 | ERCC5;YY1 | 2 | 368 | 10 | 15874 | 0.02 |
| GO:2001020 | regulation of response to DNA damage stimulus | biological_process | 7 | IER3;MCL1 | 2 | 368 | 10 | 15874 | 0.02 |
| GO:0045047 | protein targeting to ER | biological_process | 7 | SEC61G;SPCS3 | 2 | 368 | 10 | 15874 | 0.02 |
| GO:0050684 | regulation of mRNA processing | biological_process | 11 | CDK11A;PTCD2 | 2 | 368 | 10 | 15874 | 0.02 |
| GO:0015935 | small ribosomal subunit | cellular_component | 11 | RACK1;RPS16;RPS5 | 3 | 368 | 26 | 15874 | 0.02 |
| GO:0042795 | snRNA transcription from RNA polymerase II promoter | biological_process | 12 | INTS6;POLR2D;RPRD1A;SP1;TAF11 | 5 | 368 | 70 | 15874 | 0.02 |
| GO:1900102 | negative regulation of endoplasmic reticulum unfolded protein response | biological_process | 10 | RACK1 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0036517 | chemoattraction of serotonergic neuron axon | NA | NA | WNT5A | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0070377 | negative regulation of ERK5 cascade | biological_process | 12 | MAPK7 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0010009 | cytoplasmic side of endosome membrane | cellular_component | 12 | RBSN | 1 | 368 | 1 | 15874 | 0.02 |
| GO:2001145 | negative regulation of phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity | biological_process | 11 | BAG4 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0042745 | circadian sleep/wake cycle | biological_process | 6 | AHCY | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0006049 | UDP-N-acetylglucosamine catabolic process | biological_process | 11 | MGAT1 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:2000103 | positive regulation of mammary stem cell proliferation | biological_process | 6 | LBH | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0004462 | lactoylglutathione lyase activity | molecular_function | 6 | GLO1 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0050038 | L-xylulose reductase (NADP+) activity | molecular_function | 7 | DCXR | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0052835 | inositol-3,4,6-trisphosphate 1-kinase activity | molecular_function | 8 | ITPK1 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0044328 | canonical Wnt signaling pathway involved in positive regulation of endothelial cell migration | biological_process | 13 | PLPP3 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0032655 | regulation of interleukin-12 production | biological_process | 7 | HLA-B | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0042023 | DNA endoreduplication | biological_process | 11 | ZPR1 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0035838 | growing cell tip | cellular_component | 6 | PI4K2A | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0036335 | intestinal stem cell homeostasis | biological_process | 7 | LGR4 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0051343 | positive regulation of cyclic-nucleotide phosphodiesterase activity | biological_process | 7 | RACK1 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0042404 | thyroid hormone catabolic process | biological_process | 6 | DIO2 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0071629 | ubiquitin-dependent catabolism of misfolded proteins by cytoplasm-associated proteasome | biological_process | 11 | RNF126 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0090158 | endoplasmic reticulum membrane organization | biological_process | 6 | STX18 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:1904677 | positive regulation of somatic stem cell division | NA | NA | LBH | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0072202 | cell differentiation involved in metanephros development | biological_process | 8 | LGR4 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0043004 | cytoplasmic sequestering of CFTR protein | biological_process | 8 | GOPC | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0002399 | MHC class II protein complex assembly | biological_process | 10 | HLA-DMB | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0070486 | leukocyte aggregation | biological_process | 8 | SEMA4D | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0015785 | UDP-galactose transport | biological_process | 8 | SLC35A2 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:1900131 | negative regulation of lipid binding | biological_process | 7 | PEX19 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:1990261 | pre-mRNA catabolic process | biological_process | 9 | ZPR1 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:1904955 | planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation | NA | NA | WNT5A | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0072213 | metanephric capsule development | biological_process | 11 | FOXD1 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:1902379 | chemoattractant activity involved in axon guidance | molecular_function | 4 | WNT5A | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0090560 | 2-(3-amino-3-carboxypropyl)histidine synthase activity | molecular_function | 6 | DPH2 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0052725 | inositol-1,3,4-trisphosphate 6-kinase activity | molecular_function | 8 | ITPK1 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0047325 | inositol tetrakisphosphate 1-kinase activity | molecular_function | 8 | ITPK1 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0046223 | aflatoxin catabolic process | biological_process | 8 | NFE2L2 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0004638 | phosphoribosylaminoimidazole carboxylase activity | molecular_function | 7 | PAICS | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0052825 | inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity | molecular_function | 10 | ITPK1 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0015822 | ornithine transport | biological_process | 7 | SLC25A29 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0002828 | regulation of type 2 immune response | biological_process | 6 | ECM1 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:2000404 | regulation of T cell migration | biological_process | 7 | ECM1 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0036105 | peroxisome membrane class-1 targeting sequence binding | molecular_function | 9 | PEX19 | 1 | 368 | 1 | 15874 | 0.02 |
| GO:0009154 | purine ribonucleotide catabolic process | biological_process | 11 | HINT1 | 1 | 368 | 1 | 15874 | 0.02 |
document location: summary/7_differential_expression/*VS*/GO_Enrichment/*VS*_GO_enrichment_Gene.xlsx
Barplot of enriched GO terms:
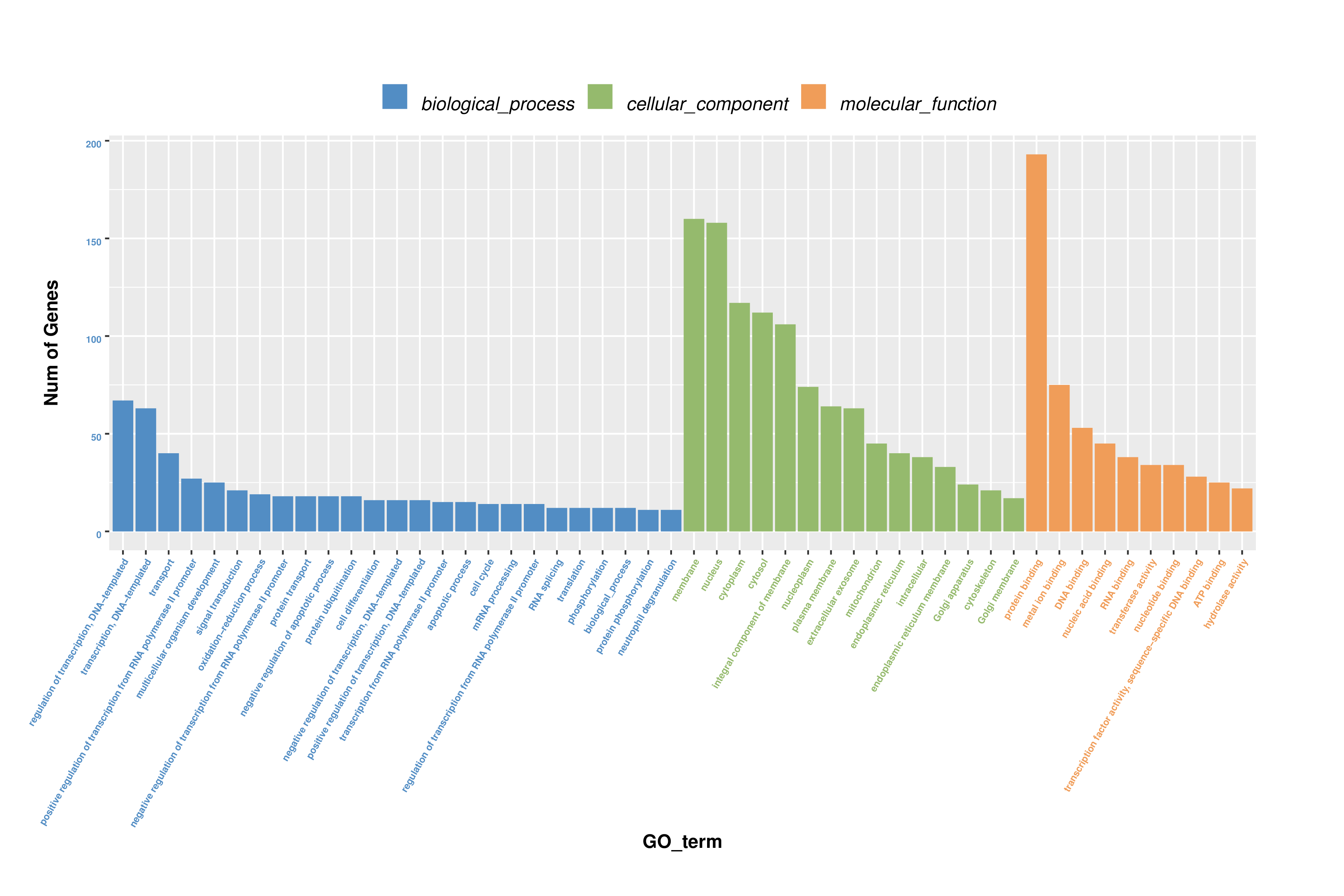
document location: summary/7_differential_expression/*VS*/1_GO_Enrichment/*VS*_GO_enrichment.png
Scatterplot of enriched GO terms:
Number of differentially expressed genes enriched in GO terms, p value and rich factor are shown in scatterplot. Rich factor=(number of differentially expressed genes in GO term)/(total number of genes in GO term). The larger rich factor is, the higher enrichment is.
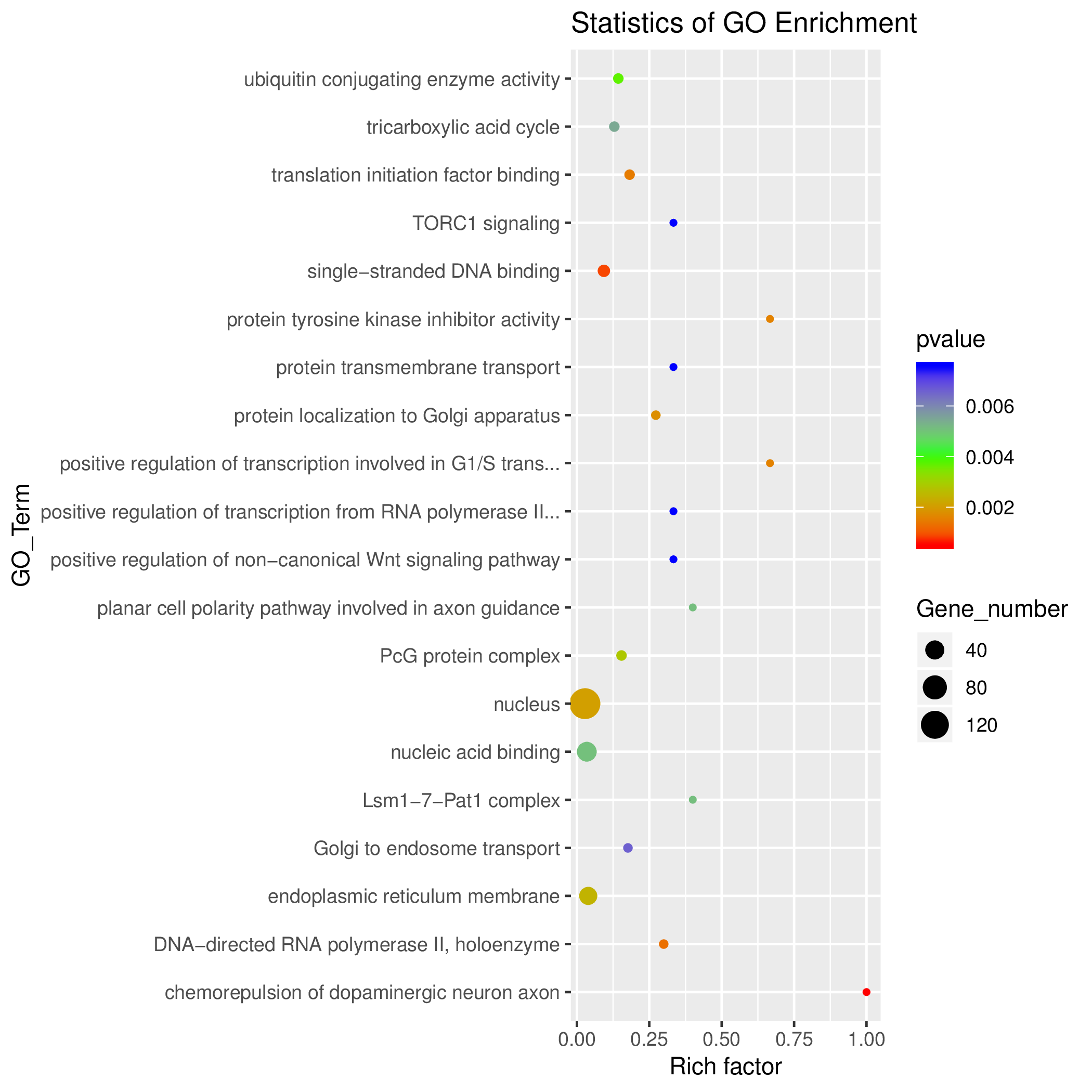
document location: summary/7_differential_expression/*VS*/1_GO_Enrichment/*VS*_GO_enrichment_scatterplot.png
KEGG (Kyoto Encyclopedia of Genes and Genomes) (http://www.kegg.jp/) is a collection of databases dealing with genomes, biological pathways, diseases, drugs, and chemical substances. KEGG is utilized for bioinformatics research and education, including data analysis in genomics, metagenomics, metabolomics and other omics studies, modeling and simulation in systems biology, and translational research in drug development.
Significant KEGG pathways were calculated by Hypergeometric equation as shown below. TB gene number=number of total genes; TS gene number=number of differentially expressed genes in total genes; B gene number=total number of genes in KEGG pathways; S gene number=number of differentially expressed genes in this KEGG pathway. Those KEGG pathways with p value<0.05 were defined as significant KEGG pathways.
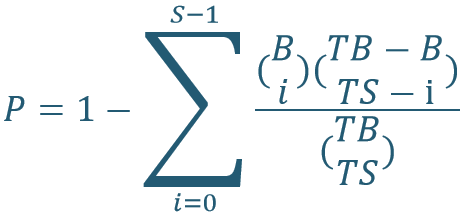
Significant KEGG pathways:
| pathway_id | pathway_name | Genes | S gene number | TS gene number | B gene number | TB gene number | pvalue |
| ko04550 | Signaling pathways regulating pluripotency of stem cells | AKT3;APC;BMI1;COMMD3-BMI1;ID2;PCGF1;PIK3R2;WNT5A;WNT5B | 9 | 131 | 122 | 6024 | 0.00 |
| ko05016 | Huntington's disease | AP2S1;CREB3;CREB5;DNAL1;NDUFA5;NDUFA7;POLR2D;SDHC;SP1;UQCRB;VDAC1 | 11 | 131 | 177 | 6024 | 0.00 |
| ko04727 | GABAergic synapse | ABAT;GABARAPL1;GABARAP;GABRE;GNG7;PRKACA | 6 | 131 | 71 | 6024 | 0.00 |
| ko00020 | Citrate cycle (TCA cycle) | CS;IDH3B;SDHC;SUCLG2 | 4 | 131 | 31 | 6024 | 0.00 |
| ko05225 | Hepatocellular carcinoma | AKT3;APC;GADD45B;IGF2;MGST3;NFE2L2;PIK3R2;WNT5A;WNT5B | 9 | 131 | 156 | 6024 | 0.01 |
| ko04145 | Phagosome | ATP6V1E1;HLA-B;HLA-DMB;SEC61G;STX18;TUBA4A;TUBB3;TUBB6 | 8 | 131 | 133 | 6024 | 0.01 |
| ko04540 | Gap junction | CSNK1D;MAPK7;PRKACA;TUBA4A;TUBB3;TUBB6 | 6 | 131 | 85 | 6024 | 0.01 |
| ko04150 | mTOR signaling pathway | AKT3;ATP6V1E1;CAB39L;LAMTOR5;MLST8;PIK3R2;WNT5A;WNT5B | 8 | 131 | 139 | 6024 | 0.01 |
| ko04137 | Mitophagy - animal | BCL2L13;CITED2;GABARAPL1;GABARAP;SP1 | 5 | 131 | 64 | 6024 | 0.01 |
| ko04962 | Vasopressin-regulated water reabsorption | CREB3;CREB5;DYNLL2;PRKACA | 4 | 131 | 42 | 6024 | 0.01 |
| ko05012 | Parkinson's disease | NDUFA5;NDUFA7;PRKACA;SDHC;UBE2J2;UQCRB;VDAC1 | 7 | 131 | 122 | 6024 | 0.02 |
| ko04915 | Estrogen signaling pathway | AKT3;CREB3;CREB5;PIK3R2;PRKACA;SP1 | 6 | 131 | 96 | 6024 | 0.02 |
| ko04725 | Cholinergic synapse | AKT3;CREB3;CREB5;GNG7;PIK3R2;PRKACA | 6 | 131 | 99 | 6024 | 0.02 |
| ko04141 | Protein processing in endoplasmic reticulum | DNAJB12;NFE2L2;SEC61G;SKP1;TXNDC5;UBE2D2;UBE2D4;UBE2J2 | 8 | 131 | 159 | 6024 | 0.02 |
| ko05224 | Breast cancer | AKT3;APC;GADD45B;PIK3R2;SP1;WNT5A;WNT5B | 7 | 131 | 133 | 6024 | 0.03 |
| ko04136 | Autophagy - other eukaryotes | GABARAPL1;GABARAP;MLST8 | 3 | 131 | 31 | 6024 | 0.03 |
| ko04927 | Cortisol synthesis and secretion | CREB3;CREB5;PRKACA;SP1 | 4 | 131 | 54 | 6024 | 0.03 |
| ko05166 | HTLV-I infection | AKT3;ANAPC11;APC;HLA-B;HLA-DMB;PIK3R2;PRKACA;VDAC1;WNT5A;WNT5B | 10 | 131 | 234 | 6024 | 0.03 |
| ko04211 | Longevity regulating pathway - mammal | AKT3;CREB3;CREB5;PIK3R2;PRKACA | 5 | 131 | 82 | 6024 | 0.03 |
| ko05217 | Basal cell carcinoma | APC;GADD45B;WNT5A;WNT5B | 4 | 131 | 57 | 6024 | 0.03 |
| ko05213 | Endometrial cancer | AKT3;APC;GADD45B;PIK3R2 | 4 | 131 | 58 | 6024 | 0.04 |
| ko04926 | Relaxin signaling pathway | AKT3;CREB3;CREB5;GNG7;PIK3R2;PRKACA | 6 | 131 | 121 | 6024 | 0.05 |
| ko04068 | FoxO signaling pathway | AKT3;GABARAPL1;GABARAP;GADD45B;PIK3R2;PLK2 | 6 | 131 | 123 | 6024 | 0.05 |
| ko04140 | Autophagy - animal | AKT3;GABARAPL1;GABARAP;MLST8;PIK3R2;PRKACA | 6 | 131 | 124 | 6024 | 0.05 |
| ko05231 | Choline metabolism in cancer | AKT3;PCYT1A;PIK3R2;PLPP3;SP1 | 5 | 131 | 95 | 6024 | 0.06 |
| ko04918 | Thyroid hormone synthesis | CREB3;CREB5;GPX2;PRKACA | 4 | 131 | 67 | 6024 | 0.06 |
| ko04371 | Apelin signaling pathway | AKT3;GABARAPL1;GABARAP;GNG7;MEF2B;PRKACA | 6 | 131 | 127 | 6024 | 0.06 |
| ko03440 | Homologous recombination | BRCC3;RPA2;XRCC2 | 3 | 131 | 41 | 6024 | 0.06 |
| ko03040 | Spliceosome | LSM2;LSM5;RBM22;RBM8A;RBMXL1;SRSF3 | 6 | 131 | 130 | 6024 | 0.06 |
| ko04210 | Apoptosis | AKT3;GADD45B;MCL1;PIK3R2;TNFRSF10B;TUBA4A | 6 | 131 | 132 | 6024 | 0.07 |
| ko04668 | TNF signaling pathway | AKT3;BAG4;CREB3;CREB5;PIK3R2 | 5 | 131 | 101 | 6024 | 0.07 |
| ko03010 | Ribosome | MRPL19;MRPL35;RPL10;RPL17;RPS16;RPS5 | 6 | 131 | 133 | 6024 | 0.07 |
| ko05210 | Colorectal cancer | AKT3;APC;GADD45B;PIK3R2 | 4 | 131 | 72 | 6024 | 0.07 |
| ko05226 | Gastric cancer | AKT3;APC;GADD45B;PIK3R2;WNT5A;WNT5B | 6 | 131 | 134 | 6024 | 0.07 |
| ko04932 | Non-alcoholic fatty liver disease (NAFLD) | AKT3;NDUFA5;NDUFA7;PIK3R2;SDHC;UQCRB | 6 | 131 | 135 | 6024 | 0.07 |
| ko03420 | Nucleotide excision repair | BIVM-ERCC5;ERCC5;RPA2 | 3 | 131 | 45 | 6024 | 0.07 |
| ko00510 | N-Glycan biosynthesis | ALG10B;DPM3;MGAT1 | 3 | 131 | 46 | 6024 | 0.08 |
| ko04923 | Regulation of lipolysis in adipocyte | AKT3;PIK3R2;PRKACA | 3 | 131 | 47 | 6024 | 0.08 |
| ko05030 | Cocaine addiction | CREB3;CREB5;PRKACA | 3 | 131 | 47 | 6024 | 0.08 |
| ko03060 | Protein export | SEC61G;SPCS3 | 2 | 131 | 23 | 6024 | 0.09 |
| ko05330 | Allograft rejection | HLA-B;HLA-DMB | 2 | 131 | 23 | 6024 | 0.09 |
| ko05110 | Vibrio cholerae infection | ATP6V1E1;PRKACA;SEC61G | 3 | 131 | 49 | 6024 | 0.09 |
| ko05206 | MicroRNAs in cancer | APC;BMI1;COMMD3-BMI1;MAPK7;MCL1;PIK3R2 | 6 | 131 | 143 | 6024 | 0.09 |
| ko04152 | AMPK signaling pathway | AKT3;CAB39L;CREB3;CREB5;PIK3R2 | 5 | 131 | 111 | 6024 | 0.09 |
| ko04919 | Thyroid hormone signaling pathway | AKT3;DIO2;MED17;PIK3R2;PRKACA | 5 | 131 | 111 | 6024 | 0.09 |
| ko04350 | TGF-beta signaling pathway | ID2;SKP1;SP1;TFDP1 | 4 | 131 | 81 | 6024 | 0.10 |
| ko05320 | Autoimmune thyroid disease | HLA-B;HLA-DMB | 2 | 131 | 25 | 6024 | 0.10 |
| ko05332 | Graft-versus-host disease | HLA-B;HLA-DMB | 2 | 131 | 25 | 6024 | 0.10 |
| ko00190 | Oxidative phosphorylation | ATP6V1E1;NDUFA5;NDUFA7;SDHC;UQCRB | 5 | 131 | 114 | 6024 | 0.10 |
| ko04218 | Cellular senescence | AKT3;GADD45B;HLA-B;PIK3R2;RBBP4;VDAC1 | 6 | 131 | 151 | 6024 | 0.11 |
| ko05130 | Pathogenic Escherichia coli infection | TUBA4A;TUBB3;TUBB6 | 3 | 131 | 55 | 6024 | 0.12 |
| ko04723 | Retrograde endocannabinoid signaling | GABRE;GNG7;NDUFA5;NDUFA7;PRKACA | 5 | 131 | 121 | 6024 | 0.12 |
| ko05202 | Transcriptional misregulation in cancers | BMI1;COMMD3-BMI1;GADD45B;H3F3A;ID2;SP1 | 6 | 131 | 156 | 6024 | 0.12 |
| ko00440 | Phosphonate and phosphinate metabolism | PCYT1A | 1 | 131 | 6 | 6024 | 0.12 |
| ko04666 | Fc gamma R-mediated phagocytosis | AKT3;CFL2;PIK3R2;PLPP3 | 4 | 131 | 88 | 6024 | 0.12 |
| ko04728 | Dopaminergic synapse | AKT3;CREB3;CREB5;GNG7;PRKACA | 5 | 131 | 122 | 6024 | 0.13 |
| ko04213 | Longevity regulating pathway - multiple species | AKT3;PIK3R2;PRKACA | 3 | 131 | 58 | 6024 | 0.13 |
| ko04914 | Progesterone-mediated oocyte maturation | AKT3;ANAPC11;PIK3R2;PRKACA | 4 | 131 | 90 | 6024 | 0.13 |
| ko05167 | Kaposi's sarcoma-associated herpesvirus infection | AKT3;GABARAPL1;GABARAP;GNG7;HLA-B;PIK3R2 | 6 | 131 | 160 | 6024 | 0.13 |
| ko04916 | Melanogenesis | CREB3;PRKACA;WNT5A;WNT5B | 4 | 131 | 91 | 6024 | 0.14 |
| ko04710 | Circadian rhythm | CSNK1D;SKP1 | 2 | 131 | 30 | 6024 | 0.14 |
| ko04940 | Type I diabetes mellitus | HLA-B;HLA-DMB | 2 | 131 | 30 | 6024 | 0.14 |
| ko01522 | Endocrine resistance | AKT3;PIK3R2;PRKACA;SP1 | 4 | 131 | 92 | 6024 | 0.14 |
| ko05161 | Hepatitis B | AKT3;CREB3;CREB5;LAMTOR5;PIK3R2 | 5 | 131 | 127 | 6024 | 0.14 |
| ko05215 | Prostate cancer | AKT3;CREB3;CREB5;PIK3R2 | 4 | 131 | 94 | 6024 | 0.15 |
| ko05418 | Fluid shear stress and atherosclerosis | AKT3;MAPK7;MGST3;NFE2L2;PIK3R2 | 5 | 131 | 129 | 6024 | 0.15 |
| ko04261 | Adrenergic signaling in cardiomyocytes | AKT3;CREB3;CREB5;PRKACA;TPM4 | 5 | 131 | 130 | 6024 | 0.15 |
| ko00640 | Propanoate metabolism | ABAT;SUCLG2 | 2 | 131 | 32 | 6024 | 0.15 |
| ko04922 | Glucagon signaling pathway | AKT3;CREB3;CREB5;PRKACA | 4 | 131 | 96 | 6024 | 0.16 |
| ko04917 | Prolactin signaling pathway | AKT3;PIK3R2;SOCS2 | 3 | 131 | 63 | 6024 | 0.16 |
| ko05031 | Amphetamine addiction | CREB3;CREB5;PRKACA | 3 | 131 | 63 | 6024 | 0.16 |
| ko04122 | Sulfur relay system | URM1 | 1 | 131 | 8 | 6024 | 0.16 |
| ko00740 | Riboflavin metabolism | ACP1 | 1 | 131 | 8 | 6024 | 0.16 |
| ko04360 | Axon guidance | CFL2;PIK3R2;RYK;SEMA4D;WNT5A;WNT5B | 6 | 131 | 169 | 6024 | 0.16 |
| ko05223 | Non-small cell lung cancer | AKT3;GADD45B;PIK3R2 | 3 | 131 | 64 | 6024 | 0.16 |
| ko04120 | Ubiquitin mediated proteolysis | ANAPC11;SKP1;UBE2D2;UBE2D4;UBE2J2 | 5 | 131 | 134 | 6024 | 0.17 |
| ko05218 | Melanoma | AKT3;GADD45B;PIK3R2 | 3 | 131 | 65 | 6024 | 0.17 |
| ko00983 | Drug metabolism - other enzymes | IMPDH1;MGST3;TK2 | 3 | 131 | 65 | 6024 | 0.17 |
| ko04310 | Wnt signaling pathway | APC;PRKACA;SKP1;WNT5A;WNT5B | 5 | 131 | 135 | 6024 | 0.17 |
| ko05165 | Human papillomavirus infection | AKT3;APC;CREB3;CREB5;HLA-B;PIK3R2;PRKACA;WNT5A;WNT5B | 9 | 131 | 287 | 6024 | 0.17 |
| ko04931 | Insulin resistance | AKT3;CREB3;CREB5;PIK3R2 | 4 | 131 | 101 | 6024 | 0.18 |
| ko01524 | Platinum drug resistance | AKT3;MGST3;PIK3R2 | 3 | 131 | 67 | 6024 | 0.18 |
| ko05214 | Glioma | AKT3;GADD45B;PIK3R2 | 3 | 131 | 68 | 6024 | 0.18 |
| ko04151 | PI3K-Akt signaling pathway | AKT3;CREB3;CREB5;GNG7;IGF2;MCL1;MLST8;PHLPP2;PIK3R2 | 9 | 131 | 294 | 6024 | 0.19 |
| ko05164 | Influenza A | AKT3;HLA-DMB;PIK3R2;TNFRSF10B;VDAC1 | 5 | 131 | 143 | 6024 | 0.20 |
| ko04390 | Hippo signaling pathway | APC;CSNK1D;ID2;WNT5A;WNT5B | 5 | 131 | 144 | 6024 | 0.20 |
| ko05034 | Alcoholism | CREB3;CREB5;GNG7;H3F3A;PRKACA | 5 | 131 | 146 | 6024 | 0.21 |
| ko05169 | Epstein-Barr virus infection | AKT3;HLA-B;PIK3R2;POLR2D;PRKACA;PSMC5 | 6 | 131 | 185 | 6024 | 0.21 |
| ko04973 | Carbohydrate digestion and absorption | AKT3;PIK3R2 | 2 | 131 | 40 | 6024 | 0.22 |
| ko05162 | Measles | AKT3;PIK3R2;RACK1;TNFRSF10B | 4 | 131 | 110 | 6024 | 0.22 |
| ko05212 | Pancreatic cancer | AKT3;GADD45B;PIK3R2 | 3 | 131 | 74 | 6024 | 0.22 |
| ko04911 | Insulin secretion | CREB3;CREB5;PRKACA | 3 | 131 | 75 | 6024 | 0.22 |
| ko05205 | Proteoglycans in cancer | AKT3;IGF2;PIK3R2;PRKACA;WNT5A;WNT5B | 6 | 131 | 188 | 6024 | 0.22 |
| ko05200 | Pathways in cancer | AKT3;APC;GADD45B;GNG7;IGF2;MGST3;NFE2L2;PIK3R2;PRKACA;SKP1;SP1;WNT5A;WNT5B | 13 | 131 | 473 | 6024 | 0.23 |
| ko05032 | Morphine addiction | GABRE;GNG7;PRKACA | 3 | 131 | 76 | 6024 | 0.23 |
| ko04022 | cGMP - PKG signaling pathway | AKT3;CREB3;CREB5;MEF2B;VDAC1 | 5 | 131 | 151 | 6024 | 0.23 |
| ko03050 | Proteasome | PSMA1;PSMC5 | 2 | 131 | 42 | 6024 | 0.23 |
| ko00730 | Thiamine metabolism | ACP1 | 1 | 131 | 12 | 6024 | 0.23 |
| ko05220 | Chronic myeloid leukemia | AKT3;GADD45B;PIK3R2 | 3 | 131 | 77 | 6024 | 0.23 |
document location: summary/7_differential_expression/*VS*/2_KEGG_Enrichment/*VS*_KEGG_enrichment_Gene.xlsx
Scatterplot of enriched KEGG pathways:
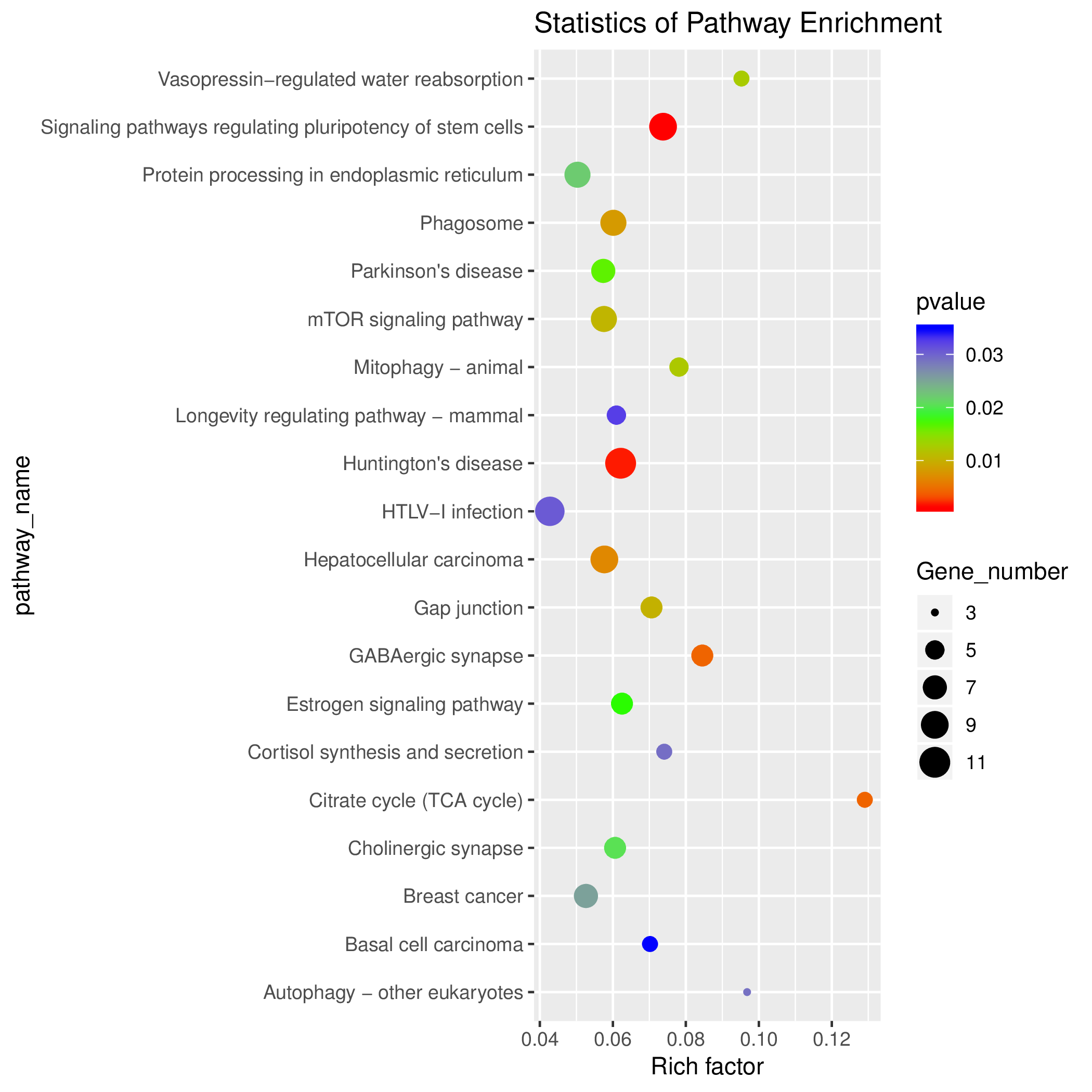
document location: summary/7_differential_expression/*VS*/2_KEGG_Enrichment/*VS*_KEGG_enrichment_scatterplot.png
KEGG pathway maps:
Graphical map objects
The KEGG pathway map is a moleculalr interaction/reaction network diagram represented in terms of the KEGG Orthology (KO) groups, so that experimental evidence in specific organisms can be generalized to other organisms through genomic information. Each map is manually drawn with in-house software called KegSketch, which generates the KGML+ file. This file is an SVG file containing graphics objects that are associated with KEGG objects (see KEGG object identifiers). Basic graphics objects in the reference KEGG pathway maps are:
boxes - ortholog (KO) groups identified by K numbers and, in metabolic maps, reactions identified by R numbers as well
circles - other molecules, usually chemical compounds identified by C numbers, but including glycans identified by G numbers
lines - reactions identified by R numbers in metabolic maps; ortholog (KO) groups identified by K numbers in global metabolism maps
and in organism specific pathway maps that are computationally generated:
boxes - genes or gene products identified by the combination of the KEGG organism code and gene identifiers
These map objects can be searched in the search box at the top of the KEGG PATHWAY page, in the search box in each pathway map, and by the KEGG Mapper tools.
Convention of map number prefix
Each pathway map is identified by the combination of 2-4 letter code and 5 digit number (see KEGG Identifiers). The prefix has the following meaning:
map - Reference pathway
ko - Reference pathway (KO)
ec - Reference pathway (EC)
rn - Reference pathway (Reaction)
org - Organism-specific pathway map
Only the first reference pathway map is manually drawn; all other maps are computationally generated. For metabolic pathways, each box (or line) in the reference map is linked to the K number (KO identifeir), the EC number, and the R number (reaction identifier). The KO, EC, and reaction maps are linked to only one of them. For all metabolic and non-metabolic maps, K numbers are converted to gene identifiers in each organism to generate organism-specific pathways.
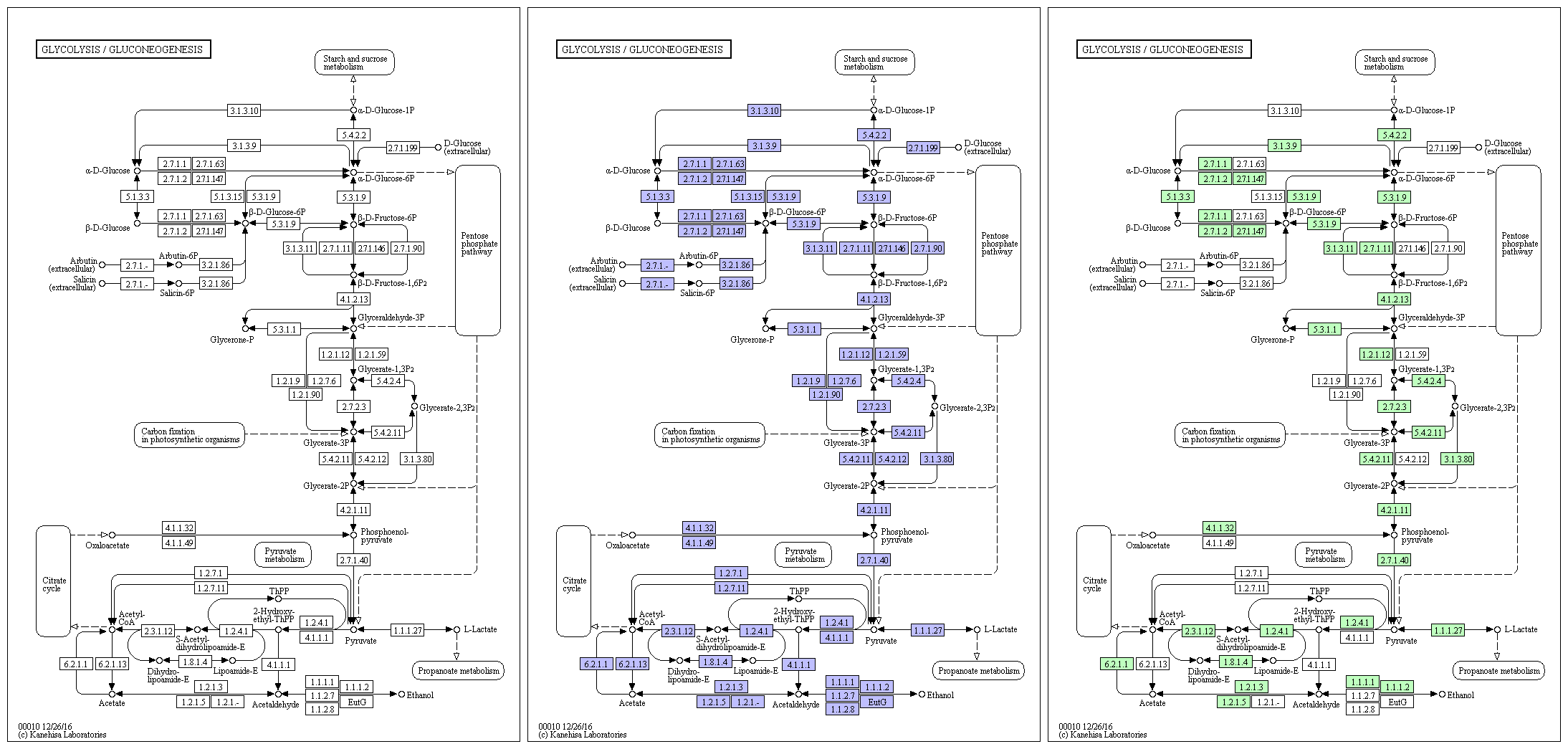
As shown above, "map" pathways are not colored, "ko/ec/rn" pathways are colored blue, and organism-specific pathways are colored green, where coloring indicates that map objects exist and are linked to corresponding entries. For global metabolism maps, "map" pathways are fully colored, so that "ko/ec/rn" pathways and organism-specific pathways are generated by reducing the coloring indicating the absence of corresponding entries.
document location: summary/7_differential_expression/*VS*/3_KEGG_Pictures
| PEAK INFO | m6A regulation INFO | gene regulation INFO | |||||||||||||||||||||
| seqnames | start | end | width | geneId | transcriptId | distanceToTSS | geneName | GO | KEGG | KO_ENTRY | EC | Description | fold_enrchment | diff.lg.fdr | diff.lg.p | diff.log2.fc | m6A_regulation | fc | log2(fc) | pval | qval | gene_regulation | significant |
| chr1 | 1046342 | 1046449 | 108 | ENSG00000188157 | ENST00000466223 | -752 | AGRN | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005829(cytosol);GO:0005509(calcium ion binding);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0007010(cytoskeleton organization);GO:0007165(signal transduction);GO:0005576(extracellular region);GO:0043236(laminin binding);GO:0007275(multicellular organism development);GO:0005578(proteinaceous extracellular matrix);GO:0030054(cell junction);GO:0045202(synapse);GO:0030154(cell differentiation);GO:0005200(structural constituent of cytoskeleton);GO:0007213(G-protein coupled acetylcholine receptor signaling pathway);GO:0043113(receptor clustering);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0070062(extracellular exosome);GO:0043547(positive regulation of GTPase activity);GO:0031012(extracellular matrix);GO:0043202(lysosomal lumen);GO:0002162(dystroglycan binding);GO:0033691(sialic acid binding);GO:0035374(chondroitin sulfate binding);GO:0043395(heparan sulfate proteoglycan binding);GO:0001523(retinoid metabolic process);GO:0006024(glycosaminoglycan biosynthetic process);GO:0006027(glycosaminoglycan catabolic process);GO:0030198(extracellular matrix organization);GO:0030203(glycosaminoglycan metabolic process);GO:0045162(clustering of voltage-gated sodium channels);GO:0045887(positive regulation of synaptic growth at neuromuscular junction);GO:0050808(synapse organization);GO:0051491(positive regulation of filopodium assembly);GO:0005605(basal lamina);GO:0005796(Golgi lumen);GO:0005796(Golgi lumen) | 04512(ECM-receptor interaction);04512(ECM-receptor interaction) | K06254 | NA | agrin [Source:HGNC Symbol;Acc:HGNC:329] | 1.58 | -1.73 | -2.96 | -1.47 | down | 1.10 | 0.14 | 0.79 | 1 | up | no |
| chr1 | 1048079 | 1048288 | 210 | ENSG00000188157 | ENST00000492947 | -241 | AGRN | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005829(cytosol);GO:0005509(calcium ion binding);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0007010(cytoskeleton organization);GO:0007165(signal transduction);GO:0005576(extracellular region);GO:0043236(laminin binding);GO:0007275(multicellular organism development);GO:0005578(proteinaceous extracellular matrix);GO:0030054(cell junction);GO:0045202(synapse);GO:0030154(cell differentiation);GO:0005200(structural constituent of cytoskeleton);GO:0007213(G-protein coupled acetylcholine receptor signaling pathway);GO:0043113(receptor clustering);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0070062(extracellular exosome);GO:0043547(positive regulation of GTPase activity);GO:0031012(extracellular matrix);GO:0043202(lysosomal lumen);GO:0002162(dystroglycan binding);GO:0033691(sialic acid binding);GO:0035374(chondroitin sulfate binding);GO:0043395(heparan sulfate proteoglycan binding);GO:0001523(retinoid metabolic process);GO:0006024(glycosaminoglycan biosynthetic process);GO:0006027(glycosaminoglycan catabolic process);GO:0030198(extracellular matrix organization);GO:0030203(glycosaminoglycan metabolic process);GO:0045162(clustering of voltage-gated sodium channels);GO:0045887(positive regulation of synaptic growth at neuromuscular junction);GO:0050808(synapse organization);GO:0051491(positive regulation of filopodium assembly);GO:0005605(basal lamina);GO:0005796(Golgi lumen);GO:0005796(Golgi lumen) | 04512(ECM-receptor interaction);04512(ECM-receptor interaction) | K06254 | NA | agrin [Source:HGNC Symbol;Acc:HGNC:329] | 1.77 | -1.56 | -2.74 | -0.44 | down | 1.10 | 0.14 | 0.79 | 1 | up | no |
| chr1 | 1055429 | 1056118 | 690 | ENSG00000242590 | ENST00000418300 | 396 | AL645608.6 | NA | NA | NA | NA | NA | 8.10 | -1.37 | -2.47 | -0.24 | down | 1 | 0 | 1 | 1 | no | no |
| chr1 | 1327709 | 1328897 | 1189 | ENSG00000169962 | ENST00000339381 | -2417 | TAS1R3 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0004930(G-protein coupled receptor activity);GO:0007186(G-protein coupled receptor signaling pathway);GO:0005886(plasma membrane);GO:0004871(signal transducer activity);GO:0007165(signal transduction);GO:0050896(response to stimulus);GO:0050909(sensory perception of taste);GO:0046982(protein heterodimerization activity);GO:0008527(taste receptor activity);GO:0033041(sweet taste receptor activity);GO:0001582(detection of chemical stimulus involved in sensory perception of sweet taste);GO:0050916(sensory perception of sweet taste);GO:0050917(sensory perception of umami taste);GO:1903767(sweet taste receptor complex) | 04973(Carbohydrate digestion and absorption);04742(Taste transduction) | K04626 | NA | taste 1 receptor member 3 [Source:HGNC Symbol;Acc:HGNC:15661] | 13.50 | -5.29 | -7.32 | -0.47 | down | 1.15 | 0.20 | 0.77 | 1 | up | no |
| chr1 | 1439706 | 1440904 | 1199 | ENSG00000179403 | ENST00000471398 | 3971 | VWA1 | GO:0005576(extracellular region);GO:0005578(proteinaceous extracellular matrix);GO:0005615(extracellular space);GO:0005604(basement membrane);GO:0070062(extracellular exosome);GO:0044267(cellular protein metabolic process);GO:0043687(post-translational protein modification);GO:0005788(endoplasmic reticulum lumen);GO:0042802(identical protein binding);GO:0030198(extracellular matrix organization);GO:0048266(behavioral response to pain);GO:0005614(interstitial matrix);GO:0005614(interstitial matrix) | NA | NA | NA | von Willebrand factor A domain containing 1 [Source:HGNC Symbol;Acc:HGNC:30910] | 14.20 | -5.65 | -7.74 | -0.35 | down | 1.01 | 0.01 | 0.95 | 1 | up | no |
| chr1 | 1534330 | 1534657 | 328 | ENSG00000205090 | ENST00000624426 | 5693 | TMEM240 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005886(plasma membrane);GO:0030054(cell junction);GO:0045202(synapse);GO:0097060(synaptic membrane);GO:0097060(synaptic membrane) | NA | NA | NA | transmembrane protein 240 [Source:HGNC Symbol;Acc:HGNC:25186] | 4.18 | -2.70 | -4.22 | -1.67 | down | 1.09 | 0.13 | 1 | 1 | up | no |
| chr1 | 2563113 | 2563352 | 240 | ENSG00000157873 | ENST00000480305 | 1312 | TNFRSF14 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0002741(positive regulation of cytokine secretion involved in immune response);GO:0046642(negative regulation of alpha-beta T cell proliferation);GO:0050731(positive regulation of peptidyl-tyrosine phosphorylation);GO:0050829(defense response to Gram-negative bacterium);GO:0050830(defense response to Gram-positive bacterium);GO:2000406(positive regulation of T cell migration);GO:0009897(external side of plasma membrane);GO:0005515(protein binding);GO:0005886(plasma membrane);GO:0005887(integral component of plasma membrane);GO:0006954(inflammatory response);GO:0046718(viral entry into host cell);GO:0097190(apoptotic signaling pathway);GO:0033209(tumor necrosis factor-mediated signaling pathway);GO:0016032(viral process);GO:0031625(ubiquitin protein ligase binding);GO:0006955(immune response);GO:0007275(multicellular organism development);GO:0032496(response to lipopolysaccharide);GO:0042127(regulation of cell proliferation);GO:0007166(cell surface receptor signaling pathway);GO:0042981(regulation of apoptotic process);GO:0001618(virus receptor activity);GO:0031295(T cell costimulation);GO:0005031(tumor necrosis factor-activated receptor activity);GO:0005031(tumor necrosis factor-activated receptor activity) | 04060(Cytokine-cytokine receptor interaction);05168(Herpes simplex infection);05168(Herpes simplex infection) | K05152 | NA | TNF receptor superfamily member 14 [Source:HGNC Symbol;Acc:HGNC:11912] | 2.92 | -2.04 | -3.36 | -1.36 | down | 0.76 | -0.40 | 0.59 | 1 | down | no |
| chr1 | 2591051 | 2591469 | 419 | ENSG00000142606 | ENST00000471840 | 1209 | MMEL1 | NA | NA | NA | NA | NA | 47.50 | -2.13 | -3.48 | -0.61 | down | 1 | 0 | 1 | 1 | no | no |
| chr1 | 6613835 | 6614104 | 270 | ENSG00000116273 | ENST00000495385 | 73 | PHF13 | GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0016569(covalent chromatin modification);GO:0051301(cell division);GO:0007049(cell cycle);GO:0005654(nucleoplasm);GO:0007059(chromosome segregation);GO:0003682(chromatin binding);GO:0030261(chromosome condensation);GO:0000278(mitotic cell cycle);GO:0035064(methylated histone binding);GO:0007076(mitotic chromosome condensation) | NA | NA | NA | PHD finger protein 13 [Source:HGNC Symbol;Acc:HGNC:22983] | 4 | -1.53 | -2.69 | -1.34 | down | 1.07 | 0.10 | 0.84 | 1 | up | no |
| chr1 | 11023310 | 11023998 | 689 | ENSG00000120948 | ENST00000617757 | 0 | TARDBP | GO:0003676(nucleic acid binding);GO:0003723(RNA binding);GO:0005634(nucleus);GO:0003730(mRNA 3'-UTR binding);GO:0006397(mRNA processing);GO:0008380(RNA splicing);GO:0005654(nucleoplasm);GO:0006366(transcription from RNA polymerase II promoter);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0005515(protein binding);GO:0001205(transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding);GO:0032024(positive regulation of insulin secretion);GO:0034976(response to endoplasmic reticulum stress);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0005726(perichromatin fibrils);GO:0005737(cytoplasm);GO:0016607(nuclear speck);GO:0035061(interchromatin granule);GO:0042802(identical protein binding);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0003690(double-stranded DNA binding);GO:0070935(3'-UTR-mediated mRNA stabilization);GO:0010629(negative regulation of gene expression);GO:0051726(regulation of cell cycle);GO:0001933(negative regulation of protein phosphorylation);GO:0042981(regulation of apoptotic process);GO:0043922(negative regulation by host of viral transcription);GO:0071765(nuclear inner membrane organization);GO:0071765(nuclear inner membrane organization) | NA | NA | NA | TAR DNA binding protein [Source:HGNC Symbol;Acc:HGNC:11571] | 4.35 | -2.38 | -3.81 | 0.33 | up | 0.99 | -0.01 | 0.97 | 1 | down | no |
| chr1 | 11750615 | 11754446 | 3832 | ENSG00000177674 | ENST00000471765 | 3103 | AGTRAP | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0043231(intracellular membrane-bounded organelle);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0005794(Golgi apparatus);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0005515(protein binding);GO:0000139(Golgi membrane);GO:0030659(cytoplasmic vesicle membrane);GO:0031410(cytoplasmic vesicle);GO:0038166(angiotensin-activated signaling pathway);GO:0004945(angiotensin type II receptor activity);GO:0001666(response to hypoxia);GO:0008217(regulation of blood pressure);GO:0005886(plasma membrane);GO:0005938(cell cortex);GO:0005938(cell cortex) | NA | NA | NA | angiotensin II receptor associated protein [Source:HGNC Symbol;Acc:HGNC:13539] | 3.80 | -1.62 | -2.81 | -1.61 | down | 1.12 | 0.17 | 0.76 | 1 | up | no |
| chr1 | 12009708 | 12011957 | 2250 | ENSG00000270914 | ENST00000603287 | 5351 | AL096840.1 | NA | NA | NA | NA | NA | 5.02 | -1.55 | -2.72 | 0.30 | up | 1 | 0 | 1 | 1 | no | no |
| chr1 | 15409895 | 15410163 | 269 | ENSG00000142634 | ENST00000375980 | 0 | EFHD2 | GO:0016020(membrane);GO:0046872(metal ion binding);GO:0005509(calcium ion binding);GO:0045121(membrane raft);GO:0045296(cadherin binding);GO:0045296(cadherin binding) | NA | NA | NA | EF-hand domain family member D2 [Source:HGNC Symbol;Acc:HGNC:28670] | 1.61 | -3.57 | -5.31 | -0.67 | down | 1.08 | 0.12 | 0.82 | 1 | up | no |
| chr1 | 15719789 | 15728093 | 8305 | ENSG00000116786 | ENST00000477849 | -1752 | PLEKHM2 | GO:0005737(cytoplasm);GO:0005515(protein binding);GO:0019894(kinesin binding);GO:0007030(Golgi organization);GO:0032418(lysosome localization);GO:0032880(regulation of protein localization);GO:0010008(endosome membrane);GO:1903527(positive regulation of membrane tubulation) | 05132(Salmonella infection) | K15348 | NA | pleckstrin homology and RUN domain containing M2 [Source:HGNC Symbol;Acc:HGNC:29131] | 13.60 | -5.08 | -7.08 | -0.51 | down | 1.03 | 0.04 | 0.91 | 1 | up | no |
| chr1 | 15784595 | 15784893 | 299 | ENSG00000162458 | ENST00000509138 | 7522 | FBLIM1 | GO:0001650(fibrillar center);GO:0005925(focal adhesion);GO:0030054(cell junction);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0007155(cell adhesion);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0005829(cytosol);GO:0001725(stress fiber);GO:0005938(cell cortex);GO:0008360(regulation of cell shape);GO:0016337(single organismal cell-cell adhesion);GO:0031005(filamin binding);GO:0033623(regulation of integrin activation);GO:0034329(cell junction assembly);GO:0034329(cell junction assembly) | NA | NA | NA | filamin binding LIM protein 1 [Source:HGNC Symbol;Acc:HGNC:24686] | 3 | -1.77 | -3.01 | -0.55 | down | 1.02 | 0.03 | 0.92 | 1 | up | no |
| chr1 | 19656869 | 19657463 | 595 | ENSG00000158748 | ENST00000289753 | -7824 | HTR6 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0004930(G-protein coupled receptor activity);GO:0007186(G-protein coupled receptor signaling pathway);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0004871(signal transducer activity);GO:0007165(signal transduction);GO:0004993(G-protein coupled serotonin receptor activity);GO:0098664(G-protein coupled serotonin receptor signaling pathway);GO:0005887(integral component of plasma membrane);GO:0007268(chemical synaptic transmission);GO:0032008(positive regulation of TOR signaling);GO:0030594(neurotransmitter receptor activity);GO:0007187(G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger);GO:0004969(histamine receptor activity);GO:0021795(cerebral cortex cell migration);GO:0005929(cilium) | 04020(Calcium signaling pathway);04024(cAMP signaling pathway);04080(Neuroactive ligand-receptor interaction);04726(Serotonergic synapse) | K04162 | NA | 5-hydroxytryptamine receptor 6 [Source:HGNC Symbol;Acc:HGNC:5301] | 30 | -2.29 | -3.70 | -0.30 | down | -inf | -inf | 1 | 1 | down | no |
| chr1 | 19656873 | 19657441 | 569 | ENSG00000158748 | ENST00000289753 | -7846 | HTR6 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0004930(G-protein coupled receptor activity);GO:0007186(G-protein coupled receptor signaling pathway);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0004871(signal transducer activity);GO:0007165(signal transduction);GO:0004993(G-protein coupled serotonin receptor activity);GO:0098664(G-protein coupled serotonin receptor signaling pathway);GO:0005887(integral component of plasma membrane);GO:0007268(chemical synaptic transmission);GO:0032008(positive regulation of TOR signaling);GO:0030594(neurotransmitter receptor activity);GO:0007187(G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger);GO:0004969(histamine receptor activity);GO:0021795(cerebral cortex cell migration);GO:0005929(cilium) | 04020(Calcium signaling pathway);04024(cAMP signaling pathway);04080(Neuroactive ligand-receptor interaction);04726(Serotonergic synapse) | K04162 | NA | 5-hydroxytryptamine receptor 6 [Source:HGNC Symbol;Acc:HGNC:5301] | 27.10 | -2.56 | -4.03 | -0.33 | down | -inf | -inf | 1 | 1 | down | no |
| chr1 | 21822198 | 21822707 | 510 | ENSG00000187942 | ENST00000484271 | 701 | LDLRAD2 | GO:0016020(membrane);GO:0016021(integral component of membrane) | NA | NA | NA | low density lipoprotein receptor class A domain containing 2 [Source:HGNC Symbol;Acc:HGNC:32071] | 24.50 | -1.45 | -2.59 | -0.64 | down | 1.09 | 0.12 | 0.81 | 1 | up | no |
| chr1 | 22864994 | 22865174 | 181 | ENSG00000133216 | ENST00000490436 | 0 | EPHB2 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0000166(nucleotide binding);GO:0004672(protein kinase activity);GO:0005524(ATP binding);GO:0006468(protein phosphorylation);GO:0005634(nucleus);GO:0016740(transferase activity);GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0004713(protein tyrosine kinase activity);GO:0005003(ephrin receptor activity);GO:0007169(transmembrane receptor protein tyrosine kinase signaling pathway);GO:0005887(integral component of plasma membrane);GO:0005886(plasma membrane);GO:0042995(cell projection);GO:0005515(protein binding);GO:0004714(transmembrane receptor protein tyrosine kinase activity);GO:0007275(multicellular organism development);GO:0007399(nervous system development);GO:0030425(dendrite);GO:0005576(extracellular region);GO:0030424(axon);GO:0005829(cytosol);GO:0010628(positive regulation of gene expression);GO:0048013(ephrin receptor signaling pathway);GO:0060997(dendritic spine morphogenesis);GO:0060021(palate development);GO:0001525(angiogenesis);GO:0007411(axon guidance);GO:0071679(commissural neuron axon guidance);GO:0051965(positive regulation of synapse assembly);GO:0042472(inner ear morphogenesis);GO:0007612(learning);GO:0007611(learning or memory);GO:0018108(peptidyl-tyrosine phosphorylation);GO:0098794(postsynapse);GO:1900273(positive regulation of long-term synaptic potentiation);GO:0001540(amyloid-beta binding);GO:0005005(transmembrane-ephrin receptor activity);GO:0044877(macromolecular complex binding);GO:0001655(urogenital system development);GO:0001933(negative regulation of protein phosphorylation);GO:0007413(axonal fasciculation);GO:0022038(corpus callosum development);GO:0031915(positive regulation of synaptic plasticity);GO:0046580(negative regulation of Ras protein signal transduction);GO:0048168(regulation of neuronal synaptic plasticity);GO:0050878(regulation of body fluid levels);GO:0051389(inactivation of MAPKK activity);GO:0060996(dendritic spine development);GO:0070373(negative regulation of ERK1 and ERK2 cascade);GO:1903078(positive regulation of protein localization to plasma membrane);GO:1904782(negative regulation of NMDA glutamate receptor activity);GO:1904783(positive regulation of NMDA glutamate receptor activity);GO:0004872(receptor activity);GO:0005102(receptor binding);GO:0008046(axon guidance receptor activity);GO:0042802(identical protein binding);GO:0000902(cell morphogenesis);GO:0009887(animal organ morphogenesis);GO:0021631(optic nerve morphogenesis);GO:0021952(central nervous system projection neuron axonogenesis);GO:0031290(retinal ganglion cell axon guidance);GO:0048170(positive regulation of long-term neuronal synaptic plasticity);GO:0048593(camera-type eye morphogenesis);GO:0050770(regulation of axonogenesis);GO:0050771(negative regulation of axonogenesis);GO:0099557(trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission);GO:0106028(-);GO:0043025(neuronal cell body);GO:0043025(neuronal cell body) | 04360(Axon guidance);04360(Axon guidance) | NA | NA | EPH receptor B2 [Source:HGNC Symbol;Acc:HGNC:3393] | 1.32 | -1.52 | -2.68 | -0.66 | down | 0.99 | -0.01 | 0.97 | 1 | down | no |
| chr1 | 24469053 | 24469263 | 211 | ENSG00000001461 | ENST00000432012 | 12887 | NIPAL3 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005515(protein binding);GO:0015095(magnesium ion transmembrane transporter activity);GO:0015693(magnesium ion transport);GO:1903830(magnesium ion transmembrane transport);GO:1903830(magnesium ion transmembrane transport) | NA | NA | NA | NIPA like domain containing 3 [Source:HGNC Symbol;Acc:HGNC:25233] | 2.20 | -2.10 | -3.44 | -1.81 | down | 0.95 | -0.07 | 0.96 | 1 | down | no |
| chr1 | 25498265 | 25498714 | 450 | ENSG00000157978 | ENST00000374338 | -44866 | LDLRAP1 | GO:0042632(cholesterol homeostasis);GO:0006810(transport);GO:0005737(cytoplasm);GO:0009967(positive regulation of signal transduction);GO:0006629(lipid metabolic process);GO:0005829(cytosol);GO:0008202(steroid metabolic process);GO:0005515(protein binding);GO:0008203(cholesterol metabolic process);GO:0005886(plasma membrane);GO:0006897(endocytosis);GO:0061024(membrane organization);GO:0005769(early endosome);GO:0009925(basal plasma membrane);GO:0048260(positive regulation of receptor-mediated endocytosis);GO:0009898(cytoplasmic side of plasma membrane);GO:0030424(axon);GO:0030665(clathrin-coated vesicle membrane);GO:0030159(receptor signaling complex scaffold activity);GO:0030276(clathrin binding);GO:0030121(AP-1 adaptor complex);GO:0035615(clathrin adaptor activity);GO:0030122(AP-2 adaptor complex);GO:0001540(amyloid-beta binding);GO:0034383(low-density lipoprotein particle clearance);GO:0090118(receptor-mediated endocytosis involved in cholesterol transport);GO:0055037(recycling endosome);GO:0050750(low-density lipoprotein particle receptor binding);GO:0006898(receptor-mediated endocytosis);GO:1905602(positive regulation of receptor-mediated endocytosis involved in cholesterol transport);GO:0001784(phosphotyrosine residue binding);GO:0090003(regulation of establishment of protein localization to plasma membrane);GO:0005883(neurofilament);GO:0005546(phosphatidylinositol-4,5-bisphosphate binding);GO:0031623(receptor internalization);GO:0043393(regulation of protein binding);GO:0035591(signaling adaptor activity);GO:0035612(AP-2 adaptor complex binding);GO:0042982(amyloid precursor protein metabolic process);GO:0090205(positive regulation of cholesterol metabolic process);GO:0090205(positive regulation of cholesterol metabolic process) | 04144(Endocytosis);04979(Cholesterol metabolism);04979(Cholesterol metabolism) | K12474 | NA | low density lipoprotein receptor adaptor protein 1 [Source:HGNC Symbol;Acc:HGNC:18640] | 4.33 | -1.76 | -3 | -1.04 | down | 1.09 | 0.13 | 0.80 | 1 | up | no |
| chr1 | 26169930 | 26170489 | 560 | ENSG00000142684 | ENST00000270812 | 0 | ZNF593 | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0008270(zinc ion binding);GO:0046872(metal ion binding);GO:0005730(nucleolus);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0005515(protein binding);GO:0030687(preribosome, large subunit precursor);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0043023(ribosomal large subunit binding);GO:0000055(ribosomal large subunit export from nucleus);GO:1903026(negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding) | NA | NA | NA | zinc finger protein 593 [Source:HGNC Symbol;Acc:HGNC:30943] | 4.95 | -3.29 | -4.97 | -0.83 | down | 1.37 | 0.45 | 0.48 | 1 | up | no |
| chr1 | 26573328 | 26575030 | 1703 | ENSG00000117676 | ENST00000438977 | 1058 | RPS6KA1 | GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0000166(nucleotide binding);GO:0004672(protein kinase activity);GO:0004674(protein serine/threonine kinase activity);GO:0005524(ATP binding);GO:0006468(protein phosphorylation);GO:0005829(cytosol);GO:0016740(transferase activity);GO:0000287(magnesium ion binding);GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0035556(intracellular signal transduction);GO:0005515(protein binding);GO:0007049(cell cycle);GO:0005654(nucleoplasm);GO:0004712(protein serine/threonine/tyrosine kinase activity);GO:0043066(negative regulation of apoptotic process);GO:0007165(signal transduction);GO:0045597(positive regulation of cell differentiation);GO:0030307(positive regulation of cell growth);GO:0043154(negative regulation of cysteine-type endopeptidase activity involved in apoptotic process);GO:0043027(cysteine-type endopeptidase inhibitor activity involved in apoptotic process);GO:0043620(regulation of DNA-templated transcription in response to stress);GO:0006915(apoptotic process);GO:0072574(hepatocyte proliferation);GO:0043555(regulation of translation in response to stress);GO:2000491(positive regulation of hepatic stellate cell activation);GO:2000491(positive regulation of hepatic stellate cell activation) | 04010(MAPK signaling pathway);04150(mTOR signaling pathway);04114(Oocyte meiosis);04914(Progesterone-mediated oocyte maturation);04720(Long-term potentiation);04722(Neurotrophin signaling pathway);04931(Insulin resistance);04931(Insulin resistance) | K04373 | EC:2.7.11.1 | ribosomal protein S6 kinase A1 [Source:HGNC Symbol;Acc:HGNC:10430] | 16.10 | -2.21 | -3.60 | -0.61 | down | 1.00 | -0.00 | 0.96 | 1 | down | no |
| chr1 | 26696040 | 26697419 | 1380 | ENSG00000117713 | ENST00000457599 | 0 | ARID1A | GO:0005634(nucleus);GO:0005654(nucleoplasm);GO:0003677(DNA binding);GO:0006338(chromatin remodeling);GO:0090544(BAF-type complex);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0016569(covalent chromatin modification);GO:0005515(protein binding);GO:0007399(nervous system development);GO:0045893(positive regulation of transcription, DNA-templated);GO:0003713(transcription coactivator activity);GO:0000790(nuclear chromatin);GO:0030521(androgen receptor signaling pathway);GO:0016922(ligand-dependent nuclear receptor binding);GO:0031491(nucleosome binding);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0001704(formation of primary germ layer);GO:0001843(neural tube closure);GO:0003205(cardiac chamber development);GO:0003408(optic cup formation involved in camera-type eye development);GO:0006325(chromatin organization);GO:0006337(nucleosome disassembly);GO:0006344(maintenance of chromatin silencing);GO:0006357(regulation of transcription from RNA polymerase II promoter);GO:0007369(gastrulation);GO:0007566(embryo implantation);GO:0019827(stem cell population maintenance);GO:0030520(intracellular estrogen receptor signaling pathway);GO:0030900(forebrain development);GO:0042766(nucleosome mobilization);GO:0042921(glucocorticoid receptor signaling pathway);GO:0043044(ATP-dependent chromatin remodeling);GO:0048096(chromatin-mediated maintenance of transcription);GO:0055007(cardiac muscle cell differentiation);GO:0060674(placenta blood vessel development);GO:1901998(toxin transport);GO:0016514(SWI/SNF complex);GO:0071564(npBAF complex);GO:0071565(nBAF complex);GO:0071565(nBAF complex) | 05225(Hepatocellular carcinoma);05225(Hepatocellular carcinoma) | K11653 | NA | AT-rich interaction domain 1A [Source:HGNC Symbol;Acc:HGNC:11110] | 18.80 | -2.02 | -3.34 | -0.72 | down | 1.09 | 0.13 | 0.80 | 1 | up | no |
| chr1 | 26774760 | 26775000 | 241 | ENSG00000117713 | ENST00000532781 | 0 | ARID1A | GO:0005634(nucleus);GO:0005654(nucleoplasm);GO:0003677(DNA binding);GO:0006338(chromatin remodeling);GO:0090544(BAF-type complex);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0016569(covalent chromatin modification);GO:0005515(protein binding);GO:0007399(nervous system development);GO:0045893(positive regulation of transcription, DNA-templated);GO:0003713(transcription coactivator activity);GO:0000790(nuclear chromatin);GO:0030521(androgen receptor signaling pathway);GO:0016922(ligand-dependent nuclear receptor binding);GO:0031491(nucleosome binding);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0001704(formation of primary germ layer);GO:0001843(neural tube closure);GO:0003205(cardiac chamber development);GO:0003408(optic cup formation involved in camera-type eye development);GO:0006325(chromatin organization);GO:0006337(nucleosome disassembly);GO:0006344(maintenance of chromatin silencing);GO:0006357(regulation of transcription from RNA polymerase II promoter);GO:0007369(gastrulation);GO:0007566(embryo implantation);GO:0019827(stem cell population maintenance);GO:0030520(intracellular estrogen receptor signaling pathway);GO:0030900(forebrain development);GO:0042766(nucleosome mobilization);GO:0042921(glucocorticoid receptor signaling pathway);GO:0043044(ATP-dependent chromatin remodeling);GO:0048096(chromatin-mediated maintenance of transcription);GO:0055007(cardiac muscle cell differentiation);GO:0060674(placenta blood vessel development);GO:1901998(toxin transport);GO:0016514(SWI/SNF complex);GO:0071564(npBAF complex);GO:0071565(nBAF complex);GO:0071565(nBAF complex) | 05225(Hepatocellular carcinoma);05225(Hepatocellular carcinoma) | K11653 | NA | AT-rich interaction domain 1A [Source:HGNC Symbol;Acc:HGNC:11110] | 1.92 | -2.03 | -3.35 | -0.88 | down | 1.09 | 0.13 | 0.80 | 1 | up | no |
| chr1 | 26863228 | 26864457 | 1230 | ENSG00000175793 | ENST00000339276 | 90 | SFN | GO:0005739(mitochondrion);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0005829(cytosol);GO:0019904(protein domain specific binding);GO:0005515(protein binding);GO:0005576(extracellular region);GO:0071901(negative regulation of protein serine/threonine kinase activity);GO:0042802(identical protein binding);GO:0006977(DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest);GO:0045296(cadherin binding);GO:0070062(extracellular exosome);GO:0061024(membrane organization);GO:0007165(signal transduction);GO:0005615(extracellular space);GO:0061436(establishment of skin barrier);GO:1900740(positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway);GO:0030659(cytoplasmic vesicle membrane);GO:0001836(release of cytochrome c from mitochondria);GO:0043154(negative regulation of cysteine-type endopeptidase activity involved in apoptotic process);GO:0045606(positive regulation of epidermal cell differentiation);GO:0008426(protein kinase C inhibitor activity);GO:0019901(protein kinase binding);GO:0051219(phosphoprotein binding);GO:0000079(regulation of cyclin-dependent protein serine/threonine kinase activity);GO:0003334(keratinocyte development);GO:0006469(negative regulation of protein kinase activity);GO:0008630(intrinsic apoptotic signaling pathway in response to DNA damage);GO:0010482(regulation of epidermal cell division);GO:0010839(negative regulation of keratinocyte proliferation);GO:0030216(keratinocyte differentiation);GO:0030307(positive regulation of cell growth);GO:0031424(keratinization);GO:0043588(skin development);GO:0046827(positive regulation of protein export from nucleus);GO:0051726(regulation of cell cycle) | 04110(Cell cycle);04115(p53 signaling pathway);04960(Aldosterone-regulated sodium reabsorption) | K06644 | NA | stratifin [Source:HGNC Symbol;Acc:HGNC:10773] | 26.10 | -3.37 | -5.06 | -0.19 | down | inf | inf | 1 | 1 | up | no |
| chr1 | 26993707 | 26994096 | 390 | ENSG00000253368 | ENST00000522111 | 0 | TRNP1 | GO:0005634(nucleus);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0007049(cell cycle);GO:0007275(multicellular organism development);GO:0007399(nervous system development);GO:0042127(regulation of cell proliferation);GO:0051726(regulation of cell cycle);GO:0005719(nuclear euchromatin);GO:0021696(cerebellar cortex morphogenesis);GO:0061351(neural precursor cell proliferation) | NA | NA | NA | TMF1-regulated nuclear protein 1 [Source:HGNC Symbol;Acc:HGNC:34348] | 3.86 | -2.99 | -4.58 | -0.50 | down | 1.19 | 0.25 | 0.67 | 1 | up | no |
| chr1 | 26994545 | 27000120 | 5576 | ENSG00000253368 | ENST00000531285 | 226 | TRNP1 | GO:0005634(nucleus);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0007049(cell cycle);GO:0007275(multicellular organism development);GO:0007399(nervous system development);GO:0042127(regulation of cell proliferation);GO:0051726(regulation of cell cycle);GO:0005719(nuclear euchromatin);GO:0021696(cerebellar cortex morphogenesis);GO:0061351(neural precursor cell proliferation) | NA | NA | NA | TMF1-regulated nuclear protein 1 [Source:HGNC Symbol;Acc:HGNC:34348] | 3.37 | -12.10 | -14.90 | -0.43 | down | 1.19 | 0.25 | 0.67 | 1 | up | no |
| chr1 | 28742979 | 28743188 | 210 | ENSG00000198492 | ENST00000468863 | 3888 | YTHDF2 | GO:0003723(RNA binding);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0005829(cytosol);GO:0000932(P-body);GO:0005515(protein binding);GO:0006959(humoral immune response);GO:1990247(N6-methyladenosine-containing RNA binding);GO:0048598(embryonic morphogenesis);GO:0043488(regulation of mRNA stability);GO:0061157(mRNA destabilization);GO:1903679(positive regulation of cap-independent translational initiation);GO:1903679(positive regulation of cap-independent translational initiation) | NA | NA | NA | YTH N6-methyladenosine RNA binding protein 2 [Source:HGNC Symbol;Acc:HGNC:31675] | 1.68 | -1.32 | -2.41 | -0.63 | down | 0.48 | -1.06 | 0.12 | 1 | down | no |
| chr1 | 31348951 | 31364392 | 15442 | ENSG00000229044 | ENST00000430143 | -2152 | AL451070.1 | NA | NA | NA | NA | NA | 3.74 | -1.72 | -2.94 | 0.57 | up | 1 | 0 | 1 | 1 | no | no |
| chr1 | 31434233 | 31434441 | 209 | ENSG00000168528 | ENST00000536384 | 20420 | SERINC2 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0006665(sphingolipid metabolic process);GO:0006658(phosphatidylserine metabolic process);GO:0070062(extracellular exosome);GO:0015194(L-serine transmembrane transporter activity);GO:0015825(L-serine transport);GO:1904219(positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity);GO:1904222(positive regulation of serine C-palmitoyltransferase activity);GO:1904222(positive regulation of serine C-palmitoyltransferase activity) | NA | NA | NA | serine incorporator 2 [Source:HGNC Symbol;Acc:HGNC:23231] | 2.11 | -2.64 | -4.15 | -0.88 | down | 0.88 | -0.18 | 0.83 | 1 | down | no |
| chr1 | 31587030 | 31587626 | 597 | ENSG00000142910 | ENST00000480586 | 1879 | TINAGL1 | GO:0005615(extracellular space);GO:0005515(protein binding);GO:0008234(cysteine-type peptidase activity);GO:0006508(proteolysis);GO:0005576(extracellular region);GO:0006955(immune response);GO:0005044(scavenger receptor activity);GO:0006898(receptor-mediated endocytosis);GO:0030247(polysaccharide binding);GO:0070062(extracellular exosome);GO:0007155(cell adhesion);GO:0016197(endosomal transport);GO:0031012(extracellular matrix);GO:0005201(extracellular matrix structural constituent);GO:0043236(laminin binding);GO:0005737(cytoplasm);GO:0005737(cytoplasm) | NA | NA | NA | tubulointerstitial nephritis antigen like 1 [Source:HGNC Symbol;Acc:HGNC:19168] | 3.02 | -2.18 | -3.56 | -0.37 | down | 0.80 | -0.32 | 0.66 | 1 | down | no |
| chr1 | 32193259 | 32195421 | 2163 | ENSG00000160050 | ENST00000373602 | -4965 | CCDC28B | GO:0005737(cytoplasm);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0005813(centrosome);GO:0005815(microtubule organizing center);GO:0060271(cilium assembly);GO:0030030(cell projection organization);GO:0030030(cell projection organization) | NA | NA | NA | coiled-coil domain containing 28B [Source:HGNC Symbol;Acc:HGNC:28163] | 9.21 | -1.38 | -2.49 | -0.29 | down | 1.16 | 0.21 | 0.72 | 1 | up | no |
| chr1 | 32770981 | 32771550 | 570 | ENSG00000162522 | ENST00000373480 | 5314 | KIAA1522 | GO:0030154(cell differentiation);GO:0030154(cell differentiation) | NA | NA | NA | KIAA1522 [Source:HGNC Symbol;Acc:HGNC:29301] | 3.67 | -3.38 | -5.07 | -0.31 | down | 1.17 | 0.23 | 0.69 | 1 | up | no |
| chr1 | 33281593 | 33281831 | 239 | ENSG00000160094 | ENST00000477934 | 1243 | ZNF362 | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0003677(DNA binding) | NA | NA | NA | zinc finger protein 362 [Source:HGNC Symbol;Acc:HGNC:18079] | 4.54 | -1.61 | -2.80 | 1.09 | up | 1.06 | 0.09 | 0.85 | 1 | up | no |
| chr1 | 36156199 | 36171546 | 15348 | ENSG00000116871 | ENST00000373151 | 0 | MAP7D1 | GO:0000226(microtubule cytoskeleton organization);GO:0015630(microtubule cytoskeleton);GO:0005737(cytoplasm);GO:0005829(cytosol);GO:0005856(cytoskeleton);GO:0005819(spindle);GO:0005198(structural molecule activity);GO:0005198(structural molecule activity) | NA | NA | NA | MAP7 domain containing 1 [Source:HGNC Symbol;Acc:HGNC:25514] | 8.10 | -2.99 | -4.58 | -0.27 | down | 0.97 | -0.05 | 0.98 | 1 | down | no |
| chr1 | 36180310 | 36180849 | 540 | ENSG00000116871 | ENST00000487114 | 1289 | MAP7D1 | GO:0000226(microtubule cytoskeleton organization);GO:0015630(microtubule cytoskeleton);GO:0005737(cytoplasm);GO:0005829(cytosol);GO:0005856(cytoskeleton);GO:0005819(spindle);GO:0005198(structural molecule activity);GO:0005198(structural molecule activity) | NA | NA | NA | MAP7 domain containing 1 [Source:HGNC Symbol;Acc:HGNC:25514] | 22.90 | -1.31 | -2.39 | -0.27 | down | 0.97 | -0.05 | 0.98 | 1 | down | no |
| chr1 | 36296667 | 36304308 | 7642 | ENSG00000214193 | ENST00000453908 | -2079 | SH3D21 | GO:0005634(nucleus);GO:0005654(nucleoplasm);GO:0005886(plasma membrane);GO:0070062(extracellular exosome);GO:0070062(extracellular exosome) | NA | NA | NA | SH3 domain containing 21 [Source:HGNC Symbol;Acc:HGNC:26236] | 23.60 | -3.44 | -5.15 | 0.48 | up | 0.73 | -0.46 | 0.54 | 1 | down | no |
| chr1 | 36322379 | 36322529 | 151 | ENSG00000142694 | ENST00000490466 | 1086 | EVA1B | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005515(protein binding);GO:0005515(protein binding) | NA | NA | NA | eva-1 homolog B [Source:HGNC Symbol;Acc:HGNC:25558] | 2.15 | -2.08 | -3.42 | -0.80 | down | 1.26 | 0.33 | 0.58 | 1 | up | no |
| chr1 | 37799584 | 37800211 | 628 | ENSG00000233728 | ENST00000433474 | 668 | AL929472.3 | NA | NA | NA | NA | NA | 3.73 | -1.68 | -2.88 | -0.29 | down | 1 | 0 | 1 | 1 | no | no |
| chr1 | 37808109 | 37808495 | 387 | ENSG00000196449 | ENST00000373044 | 0 | YRDC | GO:0016020(membrane);GO:0005739(mitochondrion);GO:0003725(double-stranded RNA binding);GO:0005737(cytoplasm);GO:0000049(tRNA binding);GO:0051051(negative regulation of transport);GO:0016779(nucleotidyltransferase activity);GO:0002949(tRNA threonylcarbamoyladenosine modification);GO:0006450(regulation of translational fidelity) | NA | NA | NA | yrdC N6-threonylcarbamoyltransferase domain containing [Source:HGNC Symbol;Acc:HGNC:28905] | 11.60 | -5.53 | -7.61 | -1.11 | down | 1.03 | 0.04 | 0.91 | 1 | up | no |
| chr1 | 39026376 | 39034636 | 8261 | ENSG00000168653 | ENST00000372969 | 58 | NDUFS5 | GO:0016020(membrane);GO:0055114(oxidation-reduction process);GO:0005743(mitochondrial inner membrane);GO:0005739(mitochondrion);GO:0070469(respiratory chain);GO:0005758(mitochondrial intermembrane space);GO:0032981(mitochondrial respiratory chain complex I assembly);GO:0008137(NADH dehydrogenase (ubiquinone) activity);GO:0006120(mitochondrial electron transport, NADH to ubiquinone);GO:0005747(mitochondrial respiratory chain complex I) | 00190(Oxidative phosphorylation);04723(Retrograde endocannabinoid signaling);05010(Alzheimer's disease);05012(Parkinson's disease);05016(Huntington's disease);04932(Non-alcoholic fatty liver disease (NAFLD)) | K03938 | NA | NADH:ubiquinone oxidoreductase subunit S5 [Source:HGNC Symbol;Acc:HGNC:7712] | 113 | -1.40 | -2.51 | -0.80 | down | 1.63 | 0.71 | 0.26 | 1 | up | no |
| chr1 | 39388387 | 39409531 | 21145 | ENSG00000127603 | ENST00000530275 | 0 | MACF1 | GO:0003779(actin binding);GO:0005509(calcium ion binding);GO:0005856(cytoskeleton);GO:0008017(microtubule binding);GO:0016055(Wnt signaling pathway);GO:0016887(ATPase activity);GO:0003723(RNA binding);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0046872(metal ion binding);GO:0005794(Golgi apparatus);GO:0005886(plasma membrane);GO:0042995(cell projection);GO:0005515(protein binding);GO:0005874(microtubule);GO:0045296(cadherin binding);GO:0051015(actin filament binding);GO:0045773(positive regulation of axon extension);GO:0032587(ruffle membrane);GO:0042060(wound healing);GO:0051893(regulation of focal adhesion assembly);GO:0032886(regulation of microtubule-based process);GO:0030177(positive regulation of Wnt signaling pathway);GO:0010632(regulation of epithelial cell migration);GO:0043001(Golgi to plasma membrane protein transport);GO:0016021(integral component of membrane);GO:0030054(cell junction);GO:0015629(actin cytoskeleton);GO:0015629(actin cytoskeleton) | NA | NA | NA | microtubule-actin crosslinking factor 1 [Source:HGNC Symbol;Acc:HGNC:13664] | 2.87 | -2.41 | -3.85 | 0.53 | up | 0.74 | -0.43 | 0.55 | 1 | down | no |
| chr1 | 39412590 | 39412830 | 241 | ENSG00000127603 | ENST00000641104 | 1567 | MACF1 | GO:0003779(actin binding);GO:0005509(calcium ion binding);GO:0005856(cytoskeleton);GO:0008017(microtubule binding);GO:0016055(Wnt signaling pathway);GO:0016887(ATPase activity);GO:0003723(RNA binding);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0046872(metal ion binding);GO:0005794(Golgi apparatus);GO:0005886(plasma membrane);GO:0042995(cell projection);GO:0005515(protein binding);GO:0005874(microtubule);GO:0045296(cadherin binding);GO:0051015(actin filament binding);GO:0045773(positive regulation of axon extension);GO:0032587(ruffle membrane);GO:0042060(wound healing);GO:0051893(regulation of focal adhesion assembly);GO:0032886(regulation of microtubule-based process);GO:0030177(positive regulation of Wnt signaling pathway);GO:0010632(regulation of epithelial cell migration);GO:0043001(Golgi to plasma membrane protein transport);GO:0016021(integral component of membrane);GO:0030054(cell junction);GO:0015629(actin cytoskeleton);GO:0015629(actin cytoskeleton) | NA | NA | NA | microtubule-actin crosslinking factor 1 [Source:HGNC Symbol;Acc:HGNC:13664] | 1.41 | -1.77 | -3.01 | 0.42 | up | 0.74 | -0.43 | 0.55 | 1 | down | no |
| chr1 | 39485708 | 39485888 | 181 | ENSG00000183682 | ENST00000331593 | -5758 | BMP8A | GO:0008083(growth factor activity);GO:0005576(extracellular region);GO:0007275(multicellular organism development);GO:0005125(cytokine activity);GO:0005615(extracellular space);GO:0030154(cell differentiation);GO:0060395(SMAD protein signal transduction);GO:0001503(ossification);GO:0051216(cartilage development);GO:0005160(transforming growth factor beta receptor binding);GO:0010862(positive regulation of pathway-restricted SMAD protein phosphorylation);GO:0030509(BMP signaling pathway);GO:0042981(regulation of apoptotic process);GO:0043408(regulation of MAPK cascade);GO:0048468(cell development);GO:2000505(regulation of energy homeostasis);GO:0046676(negative regulation of insulin secretion);GO:0070700(BMP receptor binding);GO:0002024(diet induced thermogenesis) | 04350(TGF-beta signaling pathway);04390(Hippo signaling pathway) | K16622 | NA | bone morphogenetic protein 8a [Source:HGNC Symbol;Acc:HGNC:21650] | 1.84 | -3.02 | -4.62 | 0.44 | up | 2.20 | 1.13 | 1 | 1 | up | no |
| chr1 | 43359081 | 43359533 | 453 | ENSG00000117399 | ENST00000372462 | 68 | CDC20 | GO:0005737(cytoplasm);GO:0016567(protein ubiquitination);GO:0051301(cell division);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0007049(cell cycle);GO:0007399(nervous system development);GO:0030154(cell differentiation);GO:0031145(anaphase-promoting complex-dependent catabolic process);GO:0000922(spindle pole);GO:0005815(microtubule organizing center);GO:0005654(nucleoplasm);GO:0005829(cytosol);GO:0016579(protein deubiquitination);GO:0043161(proteasome-mediated ubiquitin-dependent protein catabolic process);GO:0051437(positive regulation of ubiquitin-protein ligase activity involved in regulation of mitotic cell cycle transition);GO:0008022(protein C-terminus binding);GO:0010997(anaphase-promoting complex binding);GO:0097027(ubiquitin-protein transferase activator activity);GO:1904668(positive regulation of ubiquitin protein ligase activity);GO:0007062(sister chromatid cohesion);GO:0019899(enzyme binding);GO:0051436(negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle);GO:0042787(protein ubiquitination involved in ubiquitin-dependent protein catabolic process);GO:0051439(regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle);GO:0005680(anaphase-promoting complex);GO:0005819(spindle);GO:0090129(positive regulation of synapse maturation);GO:0031915(positive regulation of synaptic plasticity);GO:0042826(histone deacetylase binding);GO:0007064(mitotic sister chromatid cohesion);GO:0008284(positive regulation of cell proliferation);GO:0040020(regulation of meiotic nuclear division);GO:0050773(regulation of dendrite development);GO:0090307(mitotic spindle assembly);GO:0005813(centrosome);GO:0043234(protein complex);GO:0048471(perinuclear region of cytoplasm);GO:0048471(perinuclear region of cytoplasm) | 04120(Ubiquitin mediated proteolysis);04110(Cell cycle);04114(Oocyte meiosis);05203(Viral carcinogenesis);05166(HTLV-I infection);05166(HTLV-I infection) | NA | NA | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | 8.93 | -2.16 | -3.52 | -0.57 | down | 0.90 | -0.15 | 0.86 | 1 | down | no |
| chr1 | 43603627 | 43604166 | 540 | ENSG00000142949 | ENST00000463041 | -1416 | PTPRF | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0016787(hydrolase activity);GO:0007155(cell adhesion);GO:0004725(protein tyrosine phosphatase activity);GO:0016791(phosphatase activity);GO:0006470(protein dephosphorylation);GO:0016311(dephosphorylation);GO:0004721(phosphoprotein phosphatase activity);GO:0005886(plasma membrane);GO:0008201(heparin binding);GO:0070062(extracellular exosome);GO:0035335(peptidyl-tyrosine dephosphorylation);GO:0005887(integral component of plasma membrane);GO:0016477(cell migration);GO:0007185(transmembrane receptor protein tyrosine phosphatase signaling pathway);GO:0005001(transmembrane receptor protein tyrosine phosphatase activity);GO:0032403(protein complex binding);GO:0035373(chondroitin sulfate proteoglycan binding);GO:0010975(regulation of neuron projection development);GO:0031102(neuron projection regeneration);GO:0048679(regulation of axon regeneration);GO:1900121(negative regulation of receptor binding);GO:0043005(neuron projection);GO:0043025(neuronal cell body);GO:0043025(neuronal cell body) | 04514(Cell adhesion molecules (CAMs));04520(Adherens junction);04910(Insulin signaling pathway);04931(Insulin resistance);04931(Insulin resistance) | K05695 | EC:3.1.3.48 | protein tyrosine phosphatase, receptor type F [Source:HGNC Symbol;Acc:HGNC:9670] | 5.59 | -5.19 | -7.20 | -0.30 | down | 1.03 | 0.04 | 0.91 | 1 | up | no |
| chr1 | 43967603 | 43968022 | 420 | ENSG00000117408 | ENST00000486876 | 443 | IPO13 | GO:0006810(transport);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0006886(intracellular protein transport);GO:0015031(protein transport);GO:0005515(protein binding);GO:0008536(Ran GTPase binding);GO:0008139(nuclear localization sequence binding);GO:0008565(protein transporter activity);GO:0031965(nuclear membrane);GO:0006606(protein import into nucleus);GO:0006606(protein import into nucleus) | NA | NA | NA | importin 13 [Source:HGNC Symbol;Acc:HGNC:16853] | 8.32 | -1.36 | -2.45 | -1.48 | down | 0.97 | -0.05 | 0.99 | 1 | down | no |
| chr1 | 43972303 | 43972592 | 290 | ENSG00000132768 | ENST00000527319 | 834 | DPH2 | GO:0016740(transferase activity);GO:0005515(protein binding);GO:0005829(cytosol);GO:0017183(peptidyl-diphthamide biosynthetic process from peptidyl-histidine);GO:0090560(2-(3-amino-3-carboxypropyl)histidine synthase activity);GO:0090560(2-(3-amino-3-carboxypropyl)histidine synthase activity) | NA | NA | NA | DPH2 homolog [Source:HGNC Symbol;Acc:HGNC:3004] | 2.55 | -1.35 | -2.45 | -0.54 | down | 0.34 | -1.57 | 0.02 | 0.91 | down | yes |
| chr1 | 43990362 | 43990781 | 420 | ENSG00000159214 | ENST00000486064 | -578 | CCDC24 | GO:0005515(protein binding);GO:0005515(protein binding) | NA | NA | NA | coiled-coil domain containing 24 [Source:HGNC Symbol;Acc:HGNC:28688] | 2.31 | -1.47 | -2.62 | -0.23 | down | 0.84 | -0.25 | 0.74 | 1 | down | no |
| chr1 | 44632034 | 44632296 | 263 | ENSG00000187147 | ENST00000497469 | 0 | RNF220 | GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0016740(transferase activity);GO:0016567(protein ubiquitination);GO:0005515(protein binding);GO:0090263(positive regulation of canonical Wnt signaling pathway);GO:0004842(ubiquitin-protein transferase activity);GO:0051865(protein autoubiquitination);GO:0061630(ubiquitin protein ligase activity);GO:0061630(ubiquitin protein ligase activity) | NA | NA | NA | ring finger protein 220 [Source:HGNC Symbol;Acc:HGNC:25552] | 16.10 | -3.07 | -4.68 | -1.08 | down | 0.60 | -0.73 | 0.29 | 1 | down | no |
| chr1 | 45508955 | 45509194 | 240 | ENSG00000132763 | ENST00000477188 | -239 | MMACHC | GO:0005737(cytoplasm);GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0042803(protein homodimerization activity);GO:0005515(protein binding);GO:0005829(cytosol);GO:0031419(cobalamin binding);GO:0071949(FAD binding);GO:0009235(cobalamin metabolic process);GO:0043295(glutathione binding);GO:0006749(glutathione metabolic process);GO:0032451(demethylase activity);GO:0009236(cobalamin biosynthetic process);GO:0033787(cyanocobalamin reductase (cyanide-eliminating) activity);GO:0070988(demethylation);GO:0005739(mitochondrion) | 04977(Vitamin digestion and absorption) | K14618 | NA | methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria [Source:HGNC Symbol;Acc:HGNC:24525] | 2.56 | -1.46 | -2.60 | -0.70 | down | 1.03 | 0.04 | 0.91 | 1 | up | no |
| chr1 | 45606656 | 45607836 | 1181 | ENSG00000281825 | ENST00000626823 | -926 | AL355480.4 | NA | NA | NA | NA | NA | 6.35 | -3.85 | -5.64 | 0.52 | up | 1 | 0 | 1 | 1 | no | no |
| chr1 | 50970312 | 50974304 | 3993 | ENSG00000123080 | ENST00000371761 | 368 | CDKN2C | GO:0008285(negative regulation of cell proliferation);GO:0005737(cytoplasm);GO:0005634(nucleus);GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0005515(protein binding);GO:0007049(cell cycle);GO:0005829(cytosol);GO:0019901(protein kinase binding);GO:0030308(negative regulation of cell growth);GO:0007050(cell cycle arrest);GO:0000082(G1/S transition of mitotic cell cycle);GO:0004861(cyclin-dependent protein serine/threonine kinase inhibitor activity);GO:0042326(negative regulation of phosphorylation);GO:0000079(regulation of cyclin-dependent protein serine/threonine kinase activity);GO:0045736(negative regulation of cyclin-dependent protein serine/threonine kinase activity);GO:0048709(oligodendrocyte differentiation) | 04110(Cell cycle);05202(Transcriptional misregulation in cancers);05166(HTLV-I infection);01522(Endocrine resistance) | NA | NA | cyclin dependent kinase inhibitor 2C [Source:HGNC Symbol;Acc:HGNC:1789] | 4.45 | -1.56 | -2.74 | -0.48 | down | 1.34 | 0.42 | 0.49 | 1 | up | no |
| chr1 | 55064345 | 55064673 | 329 | ENSG00000162402 | ENST00000480962 | 8319 | USP24 | GO:0016579(protein deubiquitination);GO:0016787(hydrolase activity);GO:0008233(peptidase activity);GO:0008234(cysteine-type peptidase activity);GO:0006508(proteolysis);GO:0006511(ubiquitin-dependent protein catabolic process);GO:0036459(thiol-dependent ubiquitinyl hydrolase activity);GO:0005654(nucleoplasm);GO:0005654(nucleoplasm) | NA | NA | NA | ubiquitin specific peptidase 24 [Source:HGNC Symbol;Acc:HGNC:12623] | 1.83 | -2.31 | -3.72 | -0.84 | down | 1.01 | 0.01 | 0.95 | 1 | up | no |
| chr1 | 85582855 | 85583304 | 450 | ENSG00000142871 | ENST00000480413 | 1655 | CYR61 | GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0001934(positive regulation of protein phosphorylation);GO:0030335(positive regulation of cell migration);GO:0030513(positive regulation of BMP signaling pathway);GO:0070372(regulation of ERK1 and ERK2 cascade);GO:0000166(nucleotide binding);GO:0007155(cell adhesion);GO:0005576(extracellular region);GO:0005520(insulin-like growth factor binding);GO:0001558(regulation of cell growth);GO:0006935(chemotaxis);GO:0008201(heparin binding);GO:0044267(cellular protein metabolic process);GO:0031012(extracellular matrix);GO:0043687(post-translational protein modification);GO:0007267(cell-cell signaling);GO:0005578(proteinaceous extracellular matrix);GO:0045669(positive regulation of osteoblast differentiation);GO:0005788(endoplasmic reticulum lumen);GO:0009653(anatomical structure morphogenesis);GO:0008283(cell proliferation);GO:0045860(positive regulation of protein kinase activity);GO:0005178(integrin binding);GO:0019838(growth factor binding);GO:0050840(extracellular matrix binding);GO:0001649(osteoblast differentiation);GO:0002041(intussusceptive angiogenesis);GO:0003181(atrioventricular valve morphogenesis);GO:0003278(apoptotic process involved in heart morphogenesis);GO:0003281(ventricular septum development);GO:0010518(positive regulation of phospholipase activity);GO:0010811(positive regulation of cell-substrate adhesion);GO:0016337(single organismal cell-cell adhesion);GO:0030198(extracellular matrix organization);GO:0033690(positive regulation of osteoblast proliferation);GO:0043065(positive regulation of apoptotic process);GO:0043066(negative regulation of apoptotic process);GO:0043280(positive regulation of cysteine-type endopeptidase activity involved in apoptotic process);GO:0044319(wound healing, spreading of cells);GO:0045597(positive regulation of cell differentiation);GO:0060413(atrial septum morphogenesis);GO:0060548(negative regulation of cell death);GO:0060591(chondroblast differentiation);GO:0060710(chorio-allantoic fusion);GO:0060716(labyrinthine layer blood vessel development);GO:0061036(positive regulation of cartilage development);GO:0072593(reactive oxygen species metabolic process);GO:2000304(positive regulation of ceramide biosynthetic process);GO:0005615(extracellular space) | NA | NA | NA | cysteine rich angiogenic inducer 61 [Source:HGNC Symbol;Acc:HGNC:2654] | 5.26 | -1.35 | -2.45 | 0.17 | up | 0.96 | -0.06 | 0.97 | 1 | down | no |
| chr1 | 109929406 | 109930004 | 599 | ENSG00000184371 | ENST00000526001 | 13749 | CSF1 | GO:0016021(integral component of membrane);GO:0008083(growth factor activity);GO:0005125(cytokine activity);GO:0005615(extracellular space);GO:0010628(positive regulation of gene expression);GO:0048471(perinuclear region of cytoplasm);GO:0030335(positive regulation of cell migration);GO:0016020(membrane);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0005576(extracellular region);GO:0030154(cell differentiation);GO:0002376(immune system process);GO:0045087(innate immune response);GO:0006954(inflammatory response);GO:0070062(extracellular exosome);GO:0044267(cellular protein metabolic process);GO:0043687(post-translational protein modification);GO:0008283(cell proliferation);GO:0008284(positive regulation of cell proliferation);GO:0042803(protein homodimerization activity);GO:0005788(endoplasmic reticulum lumen);GO:0030097(hemopoiesis);GO:0001954(positive regulation of cell-matrix adhesion);GO:0045860(positive regulation of protein kinase activity);GO:0042117(monocyte activation);GO:0030316(osteoclast differentiation);GO:0045672(positive regulation of osteoclast differentiation);GO:0010744(positive regulation of macrophage derived foam cell differentiation);GO:0032270(positive regulation of cellular protein metabolic process);GO:0045651(positive regulation of macrophage differentiation);GO:0007169(transmembrane receptor protein tyrosine kinase signaling pathway);GO:0030225(macrophage differentiation);GO:1990682(CSF1-CSF1R complex);GO:0010759(positive regulation of macrophage chemotaxis);GO:0005157(macrophage colony-stimulating factor receptor binding);GO:0002158(osteoclast proliferation);GO:0003006(developmental process involved in reproduction);GO:0010743(regulation of macrophage derived foam cell differentiation);GO:0030278(regulation of ossification);GO:0032946(positive regulation of mononuclear cell proliferation);GO:0038145(macrophage colony-stimulating factor signaling pathway);GO:0040018(positive regulation of multicellular organism growth);GO:0042488(positive regulation of odontogenesis of dentin-containing tooth);GO:0045657(positive regulation of monocyte differentiation);GO:0046579(positive regulation of Ras protein signal transduction);GO:0048873(homeostasis of number of cells within a tissue);GO:0060444(branching involved in mammary gland duct morphogenesis);GO:0060611(mammary gland fat development);GO:0060763(mammary duct terminal end bud growth);GO:1902228(positive regulation of macrophage colony-stimulating factor signaling pathway);GO:1904141(positive regulation of microglial cell migration);GO:1904141(positive regulation of microglial cell migration) | 04014(Ras signaling pathway);04015(Rap1 signaling pathway);04010(MAPK signaling pathway);04668(TNF signaling pathway);04151(PI3K-Akt signaling pathway);04060(Cytokine-cytokine receptor interaction);04640(Hematopoietic cell lineage);04380(Osteoclast differentiation);05323(Rheumatoid arthritis);05323(Rheumatoid arthritis) | K05453 | NA | colony stimulating factor 1 [Source:HGNC Symbol;Acc:HGNC:2432] | 3.50 | -3.81 | -5.59 | -0.55 | down | 1.12 | 0.17 | 0.76 | 1 | up | no |
| chr1 | 111766908 | 111767088 | 181 | ENSG00000064703 | ENST00000534200 | 4491 | DDX20 | GO:0003676(nucleic acid binding);GO:0006397(mRNA processing);GO:0008380(RNA splicing);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0005829(cytosol);GO:0016787(hydrolase activity);GO:0004386(helicase activity);GO:0003677(DNA binding);GO:0005515(protein binding);GO:0016604(nuclear body);GO:0005730(nucleolus);GO:0000387(spliceosomal snRNP assembly);GO:0005654(nucleoplasm);GO:0010501(RNA secondary structure unwinding);GO:0043065(positive regulation of apoptotic process);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0005856(cytoskeleton);GO:0004004(ATP-dependent RNA helicase activity);GO:0051170(nuclear import);GO:0032797(SMN complex);GO:0034719(SMN-Sm protein complex);GO:0097504(Gemini of coiled bodies);GO:0006396(RNA processing);GO:0070491(repressing transcription factor binding);GO:0000244(spliceosomal tri-snRNP complex assembly);GO:0019904(protein domain specific binding);GO:0030674(protein binding, bridging);GO:0042826(histone deacetylase binding);GO:0008285(negative regulation of cell proliferation);GO:0045892(negative regulation of transcription, DNA-templated);GO:0048477(oogenesis);GO:0050810(regulation of steroid biosynthetic process);GO:0017053(transcriptional repressor complex);GO:0090571(RNA polymerase II transcription repressor complex);GO:0090571(RNA polymerase II transcription repressor complex) | 03013(RNA transport);03013(RNA transport) | K13131 | EC:3.6.4.13 | DEAD-box helicase 20 [Source:HGNC Symbol;Acc:HGNC:2743] | 2.71 | -1.38 | -2.49 | 1.99 | up | 1.02 | 0.02 | 0.93 | 1 | up | no |
| chr1 | 112456911 | 112457360 | 450 | ENSG00000143079 | ENST00000607039 | 93 | CTTNBP2NL | GO:0005737(cytoplasm);GO:0006470(protein dephosphorylation);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0015629(actin cytoskeleton);GO:0051721(protein phosphatase 2A binding);GO:0032410(negative regulation of transporter activity);GO:0034763(negative regulation of transmembrane transport);GO:0034763(negative regulation of transmembrane transport) | NA | NA | NA | CTTNBP2 N-terminal like [Source:HGNC Symbol;Acc:HGNC:25330] | 3.12 | -1.43 | -2.56 | 0.46 | up | 0.43 | -1.22 | 0.07 | 1 | down | no |
| chr1 | 147615861 | 147619524 | 3664 | ENSG00000116128 | ENST00000473292 | 4248 | BCL9 | GO:0005634(nucleus);GO:0003713(transcription coactivator activity);GO:0008013(beta-catenin binding);GO:0014908(myotube differentiation involved in skeletal muscle regeneration);GO:0035019(somatic stem cell population maintenance);GO:0035914(skeletal muscle cell differentiation);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0060070(canonical Wnt signaling pathway);GO:0005737(cytoplasm);GO:0005794(Golgi apparatus);GO:0005801(cis-Golgi network);GO:0005515(protein binding);GO:0016055(Wnt signaling pathway);GO:0005654(nucleoplasm);GO:1904837(beta-catenin-TCF complex assembly);GO:1904837(beta-catenin-TCF complex assembly) | NA | NA | NA | B-cell CLL/lymphoma 9 [Source:HGNC Symbol;Acc:HGNC:1008] | 4.68 | -1.36 | -2.46 | -0.38 | down | 1.00 | -0.00 | 0.96 | 1 | down | no |
| chr1 | 149900232 | 149900798 | 567 | ENSG00000178096 | ENST00000369150 | 606 | BOLA1 | GO:0005739(mitochondrion);GO:0005515(protein binding) | NA | NA | NA | bolA family member 1 [Source:HGNC Symbol;Acc:HGNC:24263] | 15 | -1.31 | -2.39 | -0.40 | down | 0.75 | -0.42 | 1 | 1 | down | no |
| chr1 | 150471599 | 150472347 | 749 | ENSG00000143374 | ENST00000438568 | -15017 | TARS2 | GO:0005575(cellular_component);GO:0005737(cytoplasm);GO:0006412(translation);GO:0000166(nucleotide binding);GO:0005524(ATP binding);GO:0005739(mitochondrion);GO:0042803(protein homodimerization activity);GO:0016874(ligase activity);GO:0005759(mitochondrial matrix);GO:0004812(aminoacyl-tRNA ligase activity);GO:0006418(tRNA aminoacylation for protein translation);GO:0006450(regulation of translational fidelity);GO:0016876(ligase activity, forming aminoacyl-tRNA and related compounds);GO:0043039(tRNA aminoacylation);GO:0004829(threonine-tRNA ligase activity);GO:0006435(threonyl-tRNA aminoacylation);GO:0002161(aminoacyl-tRNA editing activity);GO:0070159(mitochondrial threonyl-tRNA aminoacylation);GO:0070159(mitochondrial threonyl-tRNA aminoacylation) | 00970(Aminoacyl-tRNA biosynthesis);00970(Aminoacyl-tRNA biosynthesis) | K01868 | EC:6.1.1.3 | threonyl-tRNA synthetase 2, mitochondrial (putative) [Source:HGNC Symbol;Acc:HGNC:30740] | 4.21 | -1.44 | -2.57 | 0.33 | up | 1.05 | 0.07 | 0.88 | 1 | up | no |
| chr1 | 151286971 | 151287420 | 450 | ENSG00000143373 | ENST00000426871 | -64 | ZNF687 | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0005515(protein binding);GO:0005515(protein binding) | NA | NA | NA | zinc finger protein 687 [Source:HGNC Symbol;Acc:HGNC:29277] | 3.65 | -1.37 | -2.47 | -0.40 | down | 1.10 | 0.14 | 0.79 | 1 | up | no |
| chr1 | 151291491 | 151291731 | 241 | ENSG00000143373 | ENST00000436614 | 1040 | ZNF687 | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0005829(cytosol);GO:0005654(nucleoplasm);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0005515(protein binding);GO:0005515(protein binding) | NA | NA | NA | zinc finger protein 687 [Source:HGNC Symbol;Acc:HGNC:29277] | 8.65 | -1.85 | -3.12 | -1.87 | down | 1.10 | 0.14 | 0.79 | 1 | up | no |
| chr1 | 151759642 | 151759911 | 270 | ENSG00000143436 | ENST00000495867 | 1009 | MRPL9 | GO:0003723(RNA binding);GO:0006412(translation);GO:0005739(mitochondrion);GO:0030529(intracellular ribonucleoprotein complex);GO:0005622(intracellular);GO:0003735(structural constituent of ribosome);GO:0005840(ribosome);GO:0005515(protein binding);GO:0005743(mitochondrial inner membrane);GO:0070125(mitochondrial translational elongation);GO:0070126(mitochondrial translational termination);GO:0005761(mitochondrial ribosome);GO:0005762(mitochondrial large ribosomal subunit);GO:0005762(mitochondrial large ribosomal subunit) | 03010(Ribosome);03010(Ribosome) | K02939 | NA | mitochondrial ribosomal protein L9 [Source:HGNC Symbol;Acc:HGNC:14277] | 2.67 | -1.76 | -2.99 | 0.54 | up | 0.89 | -0.17 | 0.84 | 1 | down | no |
| chr1 | 153661237 | 153661385 | 149 | ENSG00000143553 | ENST00000478558 | 2510 | SNAPIN | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0006886(intracellular protein transport);GO:0005794(Golgi apparatus);GO:0005765(lysosomal membrane);GO:1904115(axon cytoplasm);GO:0005829(cytosol);GO:0005515(protein binding);GO:0000139(Golgi membrane);GO:0048471(perinuclear region of cytoplasm);GO:0030054(cell junction);GO:0045202(synapse);GO:0031410(cytoplasmic vesicle);GO:0005764(lysosome);GO:0031083(BLOC-1 complex);GO:0006887(exocytosis);GO:0016032(viral process);GO:0008333(endosome to lysosome transport);GO:0008089(anterograde axonal transport);GO:0032438(melanosome organization);GO:0048490(anterograde synaptic vesicle transport);GO:0030672(synaptic vesicle membrane);GO:0031175(neuron projection development);GO:0048489(synaptic vesicle transport);GO:0008021(synaptic vesicle);GO:0030141(secretory granule);GO:0007269(neurotransmitter secretion);GO:0000149(SNARE binding);GO:0007040(lysosome organization);GO:0007042(lysosomal lumen acidification);GO:0007268(chemical synaptic transmission);GO:0008090(retrograde axonal transport);GO:0010977(negative regulation of neuron projection development);GO:0016079(synaptic vesicle exocytosis);GO:0016188(synaptic vesicle maturation);GO:0031629(synaptic vesicle fusion to presynaptic active zone membrane);GO:0032418(lysosome localization);GO:0034629(cellular protein complex localization);GO:0043393(regulation of protein binding);GO:0051604(protein maturation);GO:0072553(terminal button organization);GO:0097352(autophagosome maturation);GO:1902774(late endosome to lysosome transport);GO:1902824(positive regulation of late endosome to lysosome transport);GO:0099078(BORC complex) | NA | NA | NA | SNAP associated protein [Source:HGNC Symbol;Acc:HGNC:17145] | 1.28 | -1.34 | -2.43 | -0.51 | down | 0.87 | -0.20 | 0.80 | 1 | down | no |
| chr1 | 155017775 | 155018522 | 748 | ENSG00000160685 | ENST00000292176 | 3327 | ZBTB7B | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0005654(nucleoplasm);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0005515(protein binding);GO:0007275(multicellular organism development);GO:0030154(cell differentiation);GO:0006366(transcription from RNA polymerase II promoter);GO:0007398(ectoderm development);GO:0010628(positive regulation of gene expression);GO:0000978(RNA polymerase II core promoter proximal region sequence-specific DNA binding);GO:0001077(transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0042803(protein homodimerization activity);GO:0043370(regulation of CD4-positive, alpha-beta T cell differentiation);GO:0043376(regulation of CD8-positive, alpha-beta T cell differentiation);GO:0045944(positive regulation of transcription from RNA polymerase II promoter);GO:0045944(positive regulation of transcription from RNA polymerase II promoter) | NA | NA | NA | zinc finger and BTB domain containing 7B [Source:HGNC Symbol;Acc:HGNC:18668] | 8.79 | -2.94 | -4.52 | -0.63 | down | 1.07 | 0.09 | 0.85 | 1 | up | no |
| chr1 | 155737018 | 155738530 | 1513 | ENSG00000132676 | ENST00000491777 | 6 | DAP3 | GO:0003723(RNA binding);GO:0000166(nucleotide binding);GO:0005654(nucleoplasm);GO:0005739(mitochondrion);GO:0030529(intracellular ribonucleoprotein complex);GO:0005840(ribosome);GO:0005525(GTP binding);GO:0015935(small ribosomal subunit);GO:0006915(apoptotic process);GO:0005515(protein binding);GO:0005761(mitochondrial ribosome);GO:0003735(structural constituent of ribosome);GO:0005763(mitochondrial small ribosomal subunit);GO:0005743(mitochondrial inner membrane);GO:0070125(mitochondrial translational elongation);GO:0070126(mitochondrial translational termination);GO:0097190(apoptotic signaling pathway);GO:0097190(apoptotic signaling pathway) | NA | NA | NA | death associated protein 3 [Source:HGNC Symbol;Acc:HGNC:2673] | 9.95 | -2.17 | -3.54 | 0.76 | up | 0.77 | -0.38 | 0.60 | 1 | down | no |
| chr1 | 156114674 | 156115004 | 331 | ENSG00000160789 | ENST00000368300 | 0 | LMNA | GO:0030334(regulation of cell migration);GO:0005634(nucleus);GO:0016607(nuclear speck);GO:0005198(structural molecule activity);GO:0005515(protein binding);GO:0005882(intermediate filament);GO:0005654(nucleoplasm);GO:0005829(cytosol);GO:0031012(extracellular matrix);GO:0031965(nuclear membrane);GO:0005635(nuclear envelope);GO:0036498(IRE1-mediated unfolded protein response);GO:0034504(protein localization to nucleus);GO:0071456(cellular response to hypoxia);GO:0007077(mitotic nuclear envelope disassembly);GO:0005652(nuclear lamina);GO:0007084(mitotic nuclear envelope reassembly);GO:0008285(negative regulation of cell proliferation);GO:0030951(establishment or maintenance of microtubule cytoskeleton polarity);GO:0090343(positive regulation of cell aging);GO:0006997(nucleus organization);GO:0006998(nuclear envelope organization);GO:0010628(positive regulation of gene expression);GO:0035105(sterol regulatory element binding protein import into nucleus);GO:0055015(ventricular cardiac muscle cell development);GO:0072201(negative regulation of mesenchymal cell proliferation);GO:0090201(negative regulation of release of cytochrome c from mitochondria);GO:1900180(regulation of protein localization to nucleus);GO:2001237(negative regulation of extrinsic apoptotic signaling pathway);GO:0005638(lamin filament);GO:0005638(lamin filament) | 04210(Apoptosis);05410(Hypertrophic cardiomyopathy (HCM));05412(Arrhythmogenic right ventricular cardiomyopathy (ARVC));05414(Dilated cardiomyopathy (DCM));05414(Dilated cardiomyopathy (DCM)) | K12641 | NA | lamin A/C [Source:HGNC Symbol;Acc:HGNC:6636] | 8.29 | -3.57 | -5.31 | -0.20 | down | 0.77 | -0.37 | 0.61 | 1 | down | no |
| chr1 | 156115274 | 156123200 | 7927 | ENSG00000160789 | ENST00000502357 | 0 | LMNA | GO:0030334(regulation of cell migration);GO:0005634(nucleus);GO:0016607(nuclear speck);GO:0005198(structural molecule activity);GO:0005515(protein binding);GO:0005882(intermediate filament);GO:0005654(nucleoplasm);GO:0005829(cytosol);GO:0031012(extracellular matrix);GO:0031965(nuclear membrane);GO:0005635(nuclear envelope);GO:0036498(IRE1-mediated unfolded protein response);GO:0034504(protein localization to nucleus);GO:0071456(cellular response to hypoxia);GO:0007077(mitotic nuclear envelope disassembly);GO:0005652(nuclear lamina);GO:0007084(mitotic nuclear envelope reassembly);GO:0008285(negative regulation of cell proliferation);GO:0030951(establishment or maintenance of microtubule cytoskeleton polarity);GO:0090343(positive regulation of cell aging);GO:0006997(nucleus organization);GO:0006998(nuclear envelope organization);GO:0010628(positive regulation of gene expression);GO:0035105(sterol regulatory element binding protein import into nucleus);GO:0055015(ventricular cardiac muscle cell development);GO:0072201(negative regulation of mesenchymal cell proliferation);GO:0090201(negative regulation of release of cytochrome c from mitochondria);GO:1900180(regulation of protein localization to nucleus);GO:2001237(negative regulation of extrinsic apoptotic signaling pathway);GO:0005638(lamin filament);GO:0005638(lamin filament) | 04210(Apoptosis);05410(Hypertrophic cardiomyopathy (HCM));05412(Arrhythmogenic right ventricular cardiomyopathy (ARVC));05414(Dilated cardiomyopathy (DCM));05414(Dilated cardiomyopathy (DCM)) | K12641 | NA | lamin A/C [Source:HGNC Symbol;Acc:HGNC:6636] | 1.45 | -1.48 | -2.63 | -0.44 | down | 0.77 | -0.37 | 0.61 | 1 | down | no |
| chr1 | 156641963 | 156646092 | 4130 | ENSG00000132692 | ENST00000457777 | 0 | BCAN | GO:0016020(membrane);GO:0030246(carbohydrate binding);GO:0007155(cell adhesion);GO:0005576(extracellular region);GO:0005578(proteinaceous extracellular matrix);GO:0031225(anchored component of membrane);GO:0005540(hyaluronic acid binding);GO:0043202(lysosomal lumen);GO:0030198(extracellular matrix organization);GO:0030203(glycosaminoglycan metabolic process);GO:0005796(Golgi lumen);GO:0005201(extracellular matrix structural constituent);GO:0001501(skeletal system development);GO:0007417(central nervous system development);GO:0030206(chondroitin sulfate biosynthetic process);GO:0030207(chondroitin sulfate catabolic process);GO:0030208(dermatan sulfate biosynthetic process);GO:0022617(extracellular matrix disassembly);GO:0021766(hippocampus development);GO:0021766(hippocampus development) | NA | NA | NA | brevican [Source:HGNC Symbol;Acc:HGNC:23059] | 7.08 | -3.27 | -4.94 | -0.78 | down | 1.16 | 0.22 | 0.71 | 1 | up | no |
| chr1 | 158094725 | 158095174 | 450 | ENSG00000183853 | ENST00000368172 | 8045 | KIRREL1 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0048471(perinuclear region of cytoplasm);GO:0070062(extracellular exosome);GO:0007411(axon guidance);GO:0005911(cell-cell junction);GO:0031295(T cell costimulation);GO:0017022(myosin binding);GO:0001933(negative regulation of protein phosphorylation);GO:0007588(excretion);GO:0016337(single organismal cell-cell adhesion);GO:0030838(positive regulation of actin filament polymerization);GO:0031253(cell projection membrane);GO:0043198(dendritic shaft);GO:0045121(membrane raft);GO:0045121(membrane raft) | NA | NA | NA | kirre like nephrin family adhesion molecule 1 [Source:HGNC Symbol;Acc:HGNC:15734] | 4 | -2.94 | -4.51 | -0.36 | down | 1.06 | 0.08 | 0.86 | 1 | up | no |
| chr1 | 159782219 | 159782543 | 325 | ENSG00000158716 | ENST00000368107 | 1216 | DUSP23 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0005654(nucleoplasm);GO:0016787(hydrolase activity);GO:0004725(protein tyrosine phosphatase activity);GO:0008138(protein tyrosine/serine/threonine phosphatase activity);GO:0016791(phosphatase activity);GO:0006470(protein dephosphorylation);GO:0016311(dephosphorylation);GO:0035335(peptidyl-tyrosine dephosphorylation);GO:0005829(cytosol);GO:0004721(phosphoprotein phosphatase activity);GO:0005515(protein binding);GO:0070062(extracellular exosome);GO:0004722(protein serine/threonine phosphatase activity);GO:0004722(protein serine/threonine phosphatase activity) | NA | NA | NA | dual specificity phosphatase 23 [Source:HGNC Symbol;Acc:HGNC:21480] | 34.20 | -1.64 | -2.83 | -0.35 | down | 1.03 | 0.05 | 0.90 | 1 | up | no |
| chr1 | 162383343 | 162383792 | 450 | ENSG00000239887 | ENST00000458626 | 1613 | C1orf226 | NA | NA | NA | NA | chromosome 1 open reading frame 226 [Source:HGNC Symbol;Acc:HGNC:34351] | 3.09 | -1.39 | -2.51 | 0.57 | up | 1.85 | 0.89 | 0.16 | 1 | up | no |
| chr1 | 165905929 | 165908270 | 2342 | ENSG00000283936 | ENST00000636291 | 0 | MIR3658 | NA | NA | NA | NA | NA | 3.77 | -1.73 | -2.96 | 0.43 | up | 1 | 0 | 1 | 1 | no | no |
| chr1 | 169106354 | 169106683 | 330 | ENSG00000143153 | ENST00000367815 | 31 | ATP1B1 | GO:0006810(transport);GO:0016020(membrane);GO:0006811(ion transport);GO:0006813(potassium ion transport);GO:0006814(sodium ion transport);GO:0005890(sodium:potassium-exchanging ATPase complex);GO:0005622(intracellular);GO:0016021(integral component of membrane);GO:0007155(cell adhesion);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0050900(leukocyte migration);GO:0010248(establishment or maintenance of transmembrane electrochemical gradient);GO:0090662(ATP hydrolysis coupled transmembrane transport);GO:0070062(extracellular exosome);GO:0034220(ion transmembrane transport);GO:0042383(sarcolemma);GO:1903278(positive regulation of sodium ion export from cell);GO:1903779(regulation of cardiac conduction);GO:1903561(extracellular vesicle);GO:0051117(ATPase binding);GO:0050821(protein stabilization);GO:0060048(cardiac muscle contraction);GO:0005524(ATP binding);GO:0010107(potassium ion import);GO:0014704(intercalated disc);GO:0016887(ATPase activity);GO:0023026(MHC class II protein complex binding);GO:0005391(sodium:potassium-exchanging ATPase activity);GO:0030955(potassium ion binding);GO:0031402(sodium ion binding);GO:0006883(cellular sodium ion homeostasis);GO:0030007(cellular potassium ion homeostasis);GO:0036376(sodium ion export from cell);GO:0055119(relaxation of cardiac muscle);GO:0086009(membrane repolarization);GO:0086013(membrane repolarization during cardiac muscle cell action potential);GO:0086064(cell communication by electrical coupling involved in cardiac conduction);GO:1990573(potassium ion import across plasma membrane);GO:0008144(drug binding);GO:0001671(ATPase activator activity);GO:0006874(cellular calcium ion homeostasis);GO:0010468(regulation of gene expression);GO:0010882(regulation of cardiac muscle contraction by calcium ion signaling);GO:0032781(positive regulation of ATPase activity);GO:0044861(protein transport into plasma membrane raft);GO:0046034(ATP metabolic process);GO:0072659(protein localization to plasma membrane);GO:1901018(positive regulation of potassium ion transmembrane transporter activity);GO:1903281(positive regulation of calcium:sodium antiporter activity);GO:1903288(positive regulation of potassium ion import);GO:0008022(protein C-terminus binding);GO:0019901(protein kinase binding);GO:0001666(response to hypoxia);GO:0030001(metal ion transport);GO:1903169(regulation of calcium ion transmembrane transport);GO:0005901(caveola);GO:0016323(basolateral plasma membrane);GO:0016324(apical plasma membrane);GO:0043209(myelin sheath);GO:0043209(myelin sheath) | 04024(cAMP signaling pathway);04022(cGMP - PKG signaling pathway);04911(Insulin secretion);04918(Thyroid hormone synthesis);04919(Thyroid hormone signaling pathway);04925(Aldosterone synthesis and secretion);04260(Cardiac muscle contraction);04261(Adrenergic signaling in cardiomyocytes);04970(Salivary secretion);04971(Gastric acid secretion);04972(Pancreatic secretion);04976(Bile secretion);04973(Carbohydrate digestion and absorption);04974(Protein digestion and absorption);04978(Mineral absorption);04960(Aldosterone-regulated sodium reabsorption);04961(Endocrine and other factor-regulated calcium reabsorption);04964(Proximal tubule bicarbonate reclamation);04964(Proximal tubule bicarbonate reclamation) | K01540 | NA | ATPase Na+/K+ transporting subunit beta 1 [Source:HGNC Symbol;Acc:HGNC:804] | 2.75 | -2.11 | -3.46 | -0.87 | down | 1.01 | 0.02 | 0.94 | 1 | up | no |
| chr1 | 169131346 | 169131466 | 121 | ENSG00000143153 | ENST00000494797 | 23439 | ATP1B1 | GO:0006810(transport);GO:0016020(membrane);GO:0006811(ion transport);GO:0006813(potassium ion transport);GO:0006814(sodium ion transport);GO:0005890(sodium:potassium-exchanging ATPase complex);GO:0005622(intracellular);GO:0016021(integral component of membrane);GO:0007155(cell adhesion);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0050900(leukocyte migration);GO:0010248(establishment or maintenance of transmembrane electrochemical gradient);GO:0090662(ATP hydrolysis coupled transmembrane transport);GO:0070062(extracellular exosome);GO:0034220(ion transmembrane transport);GO:0042383(sarcolemma);GO:1903278(positive regulation of sodium ion export from cell);GO:1903779(regulation of cardiac conduction);GO:1903561(extracellular vesicle);GO:0051117(ATPase binding);GO:0050821(protein stabilization);GO:0060048(cardiac muscle contraction);GO:0005524(ATP binding);GO:0010107(potassium ion import);GO:0014704(intercalated disc);GO:0016887(ATPase activity);GO:0023026(MHC class II protein complex binding);GO:0005391(sodium:potassium-exchanging ATPase activity);GO:0030955(potassium ion binding);GO:0031402(sodium ion binding);GO:0006883(cellular sodium ion homeostasis);GO:0030007(cellular potassium ion homeostasis);GO:0036376(sodium ion export from cell);GO:0055119(relaxation of cardiac muscle);GO:0086009(membrane repolarization);GO:0086013(membrane repolarization during cardiac muscle cell action potential);GO:0086064(cell communication by electrical coupling involved in cardiac conduction);GO:1990573(potassium ion import across plasma membrane);GO:0008144(drug binding);GO:0001671(ATPase activator activity);GO:0006874(cellular calcium ion homeostasis);GO:0010468(regulation of gene expression);GO:0010882(regulation of cardiac muscle contraction by calcium ion signaling);GO:0032781(positive regulation of ATPase activity);GO:0044861(protein transport into plasma membrane raft);GO:0046034(ATP metabolic process);GO:0072659(protein localization to plasma membrane);GO:1901018(positive regulation of potassium ion transmembrane transporter activity);GO:1903281(positive regulation of calcium:sodium antiporter activity);GO:1903288(positive regulation of potassium ion import);GO:0008022(protein C-terminus binding);GO:0019901(protein kinase binding);GO:0001666(response to hypoxia);GO:0030001(metal ion transport);GO:1903169(regulation of calcium ion transmembrane transport);GO:0005901(caveola);GO:0016323(basolateral plasma membrane);GO:0016324(apical plasma membrane);GO:0043209(myelin sheath);GO:0043209(myelin sheath) | 04024(cAMP signaling pathway);04022(cGMP - PKG signaling pathway);04911(Insulin secretion);04918(Thyroid hormone synthesis);04919(Thyroid hormone signaling pathway);04925(Aldosterone synthesis and secretion);04260(Cardiac muscle contraction);04261(Adrenergic signaling in cardiomyocytes);04970(Salivary secretion);04971(Gastric acid secretion);04972(Pancreatic secretion);04976(Bile secretion);04973(Carbohydrate digestion and absorption);04974(Protein digestion and absorption);04978(Mineral absorption);04960(Aldosterone-regulated sodium reabsorption);04961(Endocrine and other factor-regulated calcium reabsorption);04964(Proximal tubule bicarbonate reclamation);04964(Proximal tubule bicarbonate reclamation) | K01540 | NA | ATPase Na+/K+ transporting subunit beta 1 [Source:HGNC Symbol;Acc:HGNC:804] | 1.48 | -1.47 | -2.62 | 0.62 | up | 1.01 | 0.02 | 0.94 | 1 | up | no |
| chr1 | 171532423 | 171535458 | 3036 | ENSG00000117523 | ENST00000495585 | -6372 | PRRC2C | GO:0003723(RNA binding);GO:0016020(membrane);GO:0008022(protein C-terminus binding);GO:0002244(hematopoietic progenitor cell differentiation);GO:0002244(hematopoietic progenitor cell differentiation) | NA | NA | NA | proline rich coiled-coil 2C [Source:HGNC Symbol;Acc:HGNC:24903] | 5.25 | -10.10 | -12.70 | 0.77 | up | 0.96 | -0.06 | 0.97 | 1 | down | no |
| chr1 | 171557638 | 171558027 | 390 | ENSG00000117523 | ENST00000495585 | 15808 | PRRC2C | GO:0003723(RNA binding);GO:0016020(membrane);GO:0008022(protein C-terminus binding);GO:0002244(hematopoietic progenitor cell differentiation);GO:0002244(hematopoietic progenitor cell differentiation) | NA | NA | NA | proline rich coiled-coil 2C [Source:HGNC Symbol;Acc:HGNC:24903] | 4.08 | -3.39 | -5.09 | 0.47 | up | 0.96 | -0.06 | 0.97 | 1 | down | no |
| chr1 | 178916503 | 178916713 | 211 | ENSG00000116191 | ENST00000478871 | 22665 | RALGPS2 | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0007264(small GTPase mediated signal transduction);GO:0005886(plasma membrane);GO:0043547(positive regulation of GTPase activity);GO:0005085(guanyl-nucleotide exchange factor activity);GO:0032485(regulation of Ral protein signal transduction);GO:0005622(intracellular);GO:0005622(intracellular) | NA | NA | NA | Ral GEF with PH domain and SH3 binding motif 2 [Source:HGNC Symbol;Acc:HGNC:30279] | 2.79 | -1.64 | -2.83 | 1.43 | up | 0.80 | -0.31 | 0.67 | 1 | down | no |
| chr1 | 179917842 | 179918291 | 450 | ENSG00000143337 | ENST00000447964 | 13876 | TOR1AIP1 | GO:0005634(nucleus);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0031965(nuclear membrane);GO:0005515(protein binding);GO:0005637(nuclear inner membrane);GO:0008092(cytoskeletal protein binding);GO:0034504(protein localization to nucleus);GO:0032781(positive regulation of ATPase activity);GO:0001671(ATPase activator activity);GO:0051117(ATPase binding);GO:0005521(lamin binding);GO:0071763(nuclear membrane organization);GO:0005635(nuclear envelope);GO:0005635(nuclear envelope) | NA | NA | NA | torsin 1A interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:29456] | 3.34 | -2.62 | -4.12 | 0.57 | up | 1.14 | 0.19 | 0.73 | 1 | up | no |
| chr1 | 180196307 | 180196845 | 539 | ENSG00000116260 | ENST00000443059 | 0 | QSOX1 | GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0005794(Golgi apparatus);GO:0043231(intracellular membrane-bounded organelle);GO:0016491(oxidoreductase activity);GO:0055114(oxidation-reduction process);GO:0045454(cell redox homeostasis);GO:0045171(intercellular bridge);GO:0005576(extracellular region);GO:0000139(Golgi membrane);GO:0005615(extracellular space);GO:0003756(protein disulfide isomerase activity);GO:0016972(thiol oxidase activity);GO:0070062(extracellular exosome);GO:0044267(cellular protein metabolic process);GO:0043687(post-translational protein modification);GO:0043312(neutrophil degranulation);GO:0030173(integral component of Golgi membrane);GO:0002576(platelet degranulation);GO:0005788(endoplasmic reticulum lumen);GO:0031093(platelet alpha granule lumen);GO:0016971(flavin-linked sulfhydryl oxidase activity);GO:0016242(negative regulation of macroautophagy);GO:0035580(specific granule lumen);GO:1904724(tertiary granule lumen);GO:0005623(cell) | NA | NA | NA | quiescin sulfhydryl oxidase 1 [Source:HGNC Symbol;Acc:HGNC:9756] | 6.74 | -2.49 | -3.95 | -0.22 | down | 1.05 | 0.07 | 0.87 | 1 | up | no |
| chr1 | 200848977 | 200849246 | 270 | ENSG00000118200 | ENST00000475326 | -6848 | CAMSAP2 | GO:0005737(cytoplasm);GO:0005829(cytosol);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0031175(neuron projection development);GO:0008017(microtubule binding);GO:0005874(microtubule);GO:0005516(calmodulin binding);GO:0005813(centrosome);GO:1990752(microtubule end);GO:0000226(microtubule cytoskeleton organization);GO:0030507(spectrin binding);GO:0051011(microtubule minus-end binding);GO:0033043(regulation of organelle organization) | NA | NA | NA | calmodulin regulated spectrin associated protein family member 2 [Source:HGNC Symbol;Acc:HGNC:29188] | 2.22 | -1.42 | -2.54 | 0.92 | up | 0.82 | -0.29 | 0.69 | 1 | down | no |
| chr1 | 201782690 | 201783465 | 776 | ENSG00000134369 | ENST00000469130 | 60 | NAV1 | GO:0005737(cytoplasm);GO:0005856(cytoskeleton);GO:0007275(multicellular organism development);GO:0007399(nervous system development);GO:0030154(cell differentiation);GO:0005874(microtubule);GO:0001578(microtubule bundle formation);GO:0001764(neuron migration);GO:0015630(microtubule cytoskeleton);GO:0043194(axon initial segment);GO:0043194(axon initial segment) | NA | NA | NA | neuron navigator 1 [Source:HGNC Symbol;Acc:HGNC:15989] | 2.46 | -2.44 | -3.89 | -0.78 | down | 1.03 | 0.04 | 0.91 | 1 | up | no |
| chr1 | 201876152 | 201876421 | 270 | ENSG00000198700 | ENST00000456707 | 1837 | IPO9 | GO:0006810(transport);GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0006886(intracellular protein transport);GO:0015031(protein transport);GO:0008536(Ran GTPase binding);GO:0005515(protein binding);GO:0005829(cytosol);GO:0005635(nuclear envelope);GO:0042393(histone binding);GO:0008565(protein transporter activity);GO:0006606(protein import into nucleus);GO:0006606(protein import into nucleus) | NA | NA | NA | importin 9 [Source:HGNC Symbol;Acc:HGNC:19425] | 3.66 | -2.19 | -3.57 | 0.35 | up | 0.98 | -0.04 | 1.00 | 1 | down | no |
| chr1 | 203165520 | 203165968 | 449 | ENSG00000133055 | ENST00000621380 | 9815 | MYBPH | GO:0007155(cell adhesion);GO:0005515(protein binding);GO:0051015(actin filament binding);GO:0051371(muscle alpha-actinin binding);GO:0097493(structural molecule activity conferring elasticity);GO:0007015(actin filament organization);GO:0045214(sarcomere organization);GO:0071688(striated muscle myosin thick filament assembly);GO:0005859(muscle myosin complex);GO:0030018(Z disc);GO:0031430(M band);GO:0032982(myosin filament);GO:0008307(structural constituent of muscle);GO:0006942(regulation of striated muscle contraction) | NA | NA | NA | myosin binding protein H [Source:HGNC Symbol;Acc:HGNC:7552] | 4.93 | -3.16 | -4.80 | 0.39 | up | 3.51 | 1.81 | 1 | 1 | up | no |
| chr1 | 206684974 | 206729008 | 44035 | ENSG00000162889 | ENST00000367103 | 0 | MAPKAPK2 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0000166(nucleotide binding);GO:0004672(protein kinase activity);GO:0004674(protein serine/threonine kinase activity);GO:0005524(ATP binding);GO:0006468(protein phosphorylation);GO:0005654(nucleoplasm);GO:0016740(transferase activity);GO:0016301(kinase activity);GO:0016310(phosphorylation);GO:0005813(centrosome);GO:0005515(protein binding);GO:0006974(cellular response to DNA damage stimulus);GO:0006954(inflammatory response);GO:0005829(cytosol);GO:0070062(extracellular exosome);GO:0043488(regulation of mRNA stability);GO:0032496(response to lipopolysaccharide);GO:0005516(calmodulin binding);GO:0004871(signal transducer activity);GO:0048010(vascular endothelial growth factor receptor signaling pathway);GO:0031572(G2 DNA damage checkpoint);GO:0035924(cellular response to vascular endothelial growth factor stimulus);GO:0006950(response to stress);GO:0018105(peptidyl-serine phosphorylation);GO:1900034(regulation of cellular response to heat);GO:0000187(activation of MAPK activity);GO:0034097(response to cytokine);GO:0002224(toll-like receptor signaling pathway);GO:0007265(Ras protein signal transduction);GO:0032680(regulation of tumor necrosis factor production);GO:0070935(3'-UTR-mediated mRNA stabilization);GO:0006691(leukotriene metabolic process);GO:0000165(MAPK cascade);GO:0004683(calmodulin-dependent protein kinase activity);GO:0009931(calcium-dependent protein serine/threonine kinase activity);GO:0051019(mitogen-activated protein kinase binding);GO:0032675(regulation of interleukin-6 production);GO:0044351(macropinocytosis);GO:0046777(protein autophosphorylation);GO:0038066(p38MAPK cascade);GO:0042535(positive regulation of tumor necrosis factor biosynthetic process);GO:0048255(mRNA stabilization);GO:0048839(inner ear development);GO:0048839(inner ear development) | 04010(MAPK signaling pathway);04370(VEGF signaling pathway);04218(Cellular senescence);04722(Neurotrophin signaling pathway);05203(Viral carcinogenesis);05167(Kaposi's sarcoma-associated herpesvirus infection);05167(Kaposi's sarcoma-associated herpesvirus infection) | K04443 | EC:2.7.11.1 | mitogen-activated protein kinase-activated protein kinase 2 [Source:HGNC Symbol;Acc:HGNC:6887] | 8.76 | -1.38 | -2.49 | -0.32 | down | 1.04 | 0.05 | 0.89 | 1 | up | no |
| chr1 | 212444344 | 212446379 | 2036 | ENSG00000117691 | ENST00000472389 | 11353 | NENF | GO:0046872(metal ion binding);GO:0005576(extracellular region);GO:0005615(extracellular space);GO:0012505(endomembrane system);GO:0016020(membrane);GO:0032099(negative regulation of appetite);GO:0008083(growth factor activity);GO:0043410(positive regulation of MAPK cascade);GO:0043410(positive regulation of MAPK cascade) | NA | NA | NA | neudesin neurotrophic factor [Source:HGNC Symbol;Acc:HGNC:30384] | 14.20 | -1.35 | -2.45 | -0.39 | down | 1.13 | 0.18 | 0.75 | 1 | up | no |
| chr1 | 214663689 | 214663869 | 181 | ENSG00000117724 | ENST00000469862 | 8541 | CENPF | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0015031(protein transport);GO:0005654(nucleoplasm);GO:0051301(cell division);GO:0005694(chromosome);GO:0042803(protein homodimerization activity);GO:0008134(transcription factor binding);GO:0005856(cytoskeleton);GO:0005515(protein binding);GO:0007049(cell cycle);GO:0007275(multicellular organism development);GO:0048471(perinuclear region of cytoplasm);GO:0030154(cell differentiation);GO:0000922(spindle pole);GO:0005819(spindle);GO:0030496(midbody);GO:0007059(chromosome segregation);GO:0000775(chromosome, centromeric region);GO:0000776(kinetochore);GO:0000777(condensed chromosome kinetochore);GO:0005829(cytosol);GO:0003682(chromatin binding);GO:0008283(cell proliferation);GO:0070840(dynein complex binding);GO:0007517(muscle organ development);GO:0008022(protein C-terminus binding);GO:0071897(DNA biosynthetic process);GO:0007062(sister chromatid cohesion);GO:0045892(negative regulation of transcription, DNA-templated);GO:0000785(chromatin);GO:0016363(nuclear matrix);GO:0005635(nuclear envelope);GO:0000278(mitotic cell cycle);GO:0051310(metaphase plate congression);GO:0007094(mitotic spindle assembly checkpoint);GO:0000940(condensed chromosome outer kinetochore);GO:0042493(response to drug);GO:0051726(regulation of cell cycle);GO:0016202(regulation of striated muscle tissue development);GO:0001822(kidney development);GO:0010389(regulation of G2/M transition of mitotic cell cycle);GO:0021591(ventricular system development);GO:0051382(kinetochore assembly);GO:0005813(centrosome);GO:0005930(axoneme);GO:0036064(ciliary basal body);GO:0045120(pronucleus);GO:0097539(ciliary transition fiber);GO:0097539(ciliary transition fiber) | NA | NA | NA | centromere protein F [Source:HGNC Symbol;Acc:HGNC:1857] | 1.97 | -3.14 | -4.78 | 1.82 | up | 0.88 | -0.18 | 0.82 | 1 | down | no |
| chr1 | 215620254 | 215620493 | 240 | ENSG00000136636 | ENST00000465650 | 1549 | KCTD3 | GO:0051260(protein homooligomerization);GO:0051260(protein homooligomerization) | NA | NA | NA | potassium channel tetramerization domain containing 3 [Source:HGNC Symbol;Acc:HGNC:21305] | 2.17 | -2.45 | -3.90 | 0.78 | up | 0.89 | -0.16 | 0.85 | 1 | down | no |
| chr1 | 223752923 | 223757375 | 4453 | ENSG00000162909 | ENST00000474026 | -249 | CAPN2 | GO:0016787(hydrolase activity);GO:0005622(intracellular);GO:0008233(peptidase activity);GO:0008234(cysteine-type peptidase activity);GO:0006508(proteolysis);GO:0004198(calcium-dependent cysteine-type endopeptidase activity);GO:0005737(cytoplasm);GO:0016020(membrane);GO:0046872(metal ion binding);GO:0005829(cytosol);GO:0005509(calcium ion binding);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0005925(focal adhesion);GO:0030425(dendrite);GO:0051493(regulation of cytoskeleton organization);GO:0071230(cellular response to amino acid stimulus);GO:0070062(extracellular exosome);GO:0005783(endoplasmic reticulum);GO:0022617(extracellular matrix disassembly);GO:0045121(membrane raft);GO:0005794(Golgi apparatus);GO:0008092(cytoskeletal protein binding);GO:0051603(proteolysis involved in cellular protein catabolic process);GO:0046982(protein heterodimerization activity);GO:0001666(response to hypoxia);GO:0001824(blastocyst development);GO:0007520(myoblast fusion);GO:0016540(protein autoprocessing);GO:0000785(chromatin);GO:0005634(nucleus);GO:0005764(lysosome);GO:0030864(cortical actin cytoskeleton);GO:0031143(pseudopodium);GO:0097038(perinuclear endoplasmic reticulum);GO:0097038(perinuclear endoplasmic reticulum) | 04141(Protein processing in endoplasmic reticulum);04210(Apoptosis);04217(Necroptosis);04218(Cellular senescence);04510(Focal adhesion);05010(Alzheimer's disease);05010(Alzheimer's disease) | K03853 | EC:3.4.22.53 | calpain 2 [Source:HGNC Symbol;Acc:HGNC:1479] | 1.59 | -1.85 | -3.12 | 0.26 | up | 0.99 | -0.02 | 0.98 | 1 | down | no |
| chr1 | 231000180 | 231000536 | 357 | ENSG00000173409 | ENST00000459891 | 4420 | ARV1 | GO:0006810(transport);GO:0016020(membrane);GO:0016021(integral component of membrane);GO:0006629(lipid metabolic process);GO:0005783(endoplasmic reticulum);GO:0005789(endoplasmic reticulum membrane);GO:0008202(steroid metabolic process);GO:0006869(lipid transport);GO:0008206(bile acid metabolic process);GO:0030301(cholesterol transport);GO:0090181(regulation of cholesterol metabolic process);GO:0008203(cholesterol metabolic process);GO:0005794(Golgi apparatus);GO:0006695(cholesterol biosynthetic process);GO:0015248(sterol transporter activity);GO:0032383(regulation of intracellular cholesterol transport);GO:0006665(sphingolipid metabolic process);GO:0097036(regulation of plasma membrane sterol distribution);GO:0032541(cortical endoplasmic reticulum);GO:0032541(cortical endoplasmic reticulum) | NA | NA | NA | ARV1 homolog, fatty acid homeostasis modulator [Source:HGNC Symbol;Acc:HGNC:29561] | 9.31 | -1.35 | -2.45 | -0.59 | down | 1.08 | 0.11 | 0.83 | 1 | up | no |
| chr1 | 234607008 | 234609483 | 2476 | ENSG00000168264 | ENST00000491430 | 0 | IRF2BP2 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0046872(metal ion binding);GO:0005654(nucleoplasm);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated) | NA | NA | NA | interferon regulatory factor 2 binding protein 2 [Source:HGNC Symbol;Acc:HGNC:21729] | 3.07 | -1.36 | -2.46 | 0.16 | up | 1.07 | 0.10 | 0.83 | 1 | up | no |
| chr1 | 244053839 | 244054435 | 597 | ENSG00000179456 | ENST00000358704 | 2556 | ZBTB18 | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0005654(nucleoplasm);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0045171(intercellular bridge);GO:0015630(microtubule cytoskeleton);GO:0005515(protein binding);GO:0007275(multicellular organism development);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0043565(sequence-specific DNA binding);GO:0045892(negative regulation of transcription, DNA-templated);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0007519(skeletal muscle tissue development);GO:0000228(nuclear chromosome);GO:0000228(nuclear chromosome) | NA | NA | NA | zinc finger and BTB domain containing 18 [Source:HGNC Symbol;Acc:HGNC:13030] | 2.38 | -4.73 | -6.67 | 0.47 | up | 0.93 | -0.10 | 0.92 | 1 | down | no |
| chr1 | 244054555 | 244055389 | 835 | ENSG00000179456 | ENST00000358704 | 3272 | ZBTB18 | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0005654(nucleoplasm);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0045171(intercellular bridge);GO:0015630(microtubule cytoskeleton);GO:0005515(protein binding);GO:0007275(multicellular organism development);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0043565(sequence-specific DNA binding);GO:0045892(negative regulation of transcription, DNA-templated);GO:0003700(transcription factor activity, sequence-specific DNA binding);GO:0007519(skeletal muscle tissue development);GO:0000228(nuclear chromosome);GO:0000228(nuclear chromosome) | NA | NA | NA | zinc finger and BTB domain containing 18 [Source:HGNC Symbol;Acc:HGNC:13030] | 5.83 | -2.44 | -3.89 | 0.27 | up | 0.93 | -0.10 | 0.92 | 1 | down | no |
| chr1 | 248847393 | 248847961 | 569 | ENSG00000171163 | ENST00000462037 | 6562 | ZNF692 | GO:0003676(nucleic acid binding);GO:0005634(nucleus);GO:0046872(metal ion binding);GO:0005730(nucleolus);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0003677(DNA binding);GO:0000978(RNA polymerase II core promoter proximal region sequence-specific DNA binding);GO:0001078(transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding);GO:0000122(negative regulation of transcription from RNA polymerase II promoter);GO:0000122(negative regulation of transcription from RNA polymerase II promoter) | NA | NA | NA | zinc finger protein 692 [Source:HGNC Symbol;Acc:HGNC:26049] | 1.76 | -3.20 | -4.86 | -0.52 | down | 0.77 | -0.37 | 0.61 | 1 | down | no |
| chr1 | 975204 | 975412 | 209 | ENSG00000187642 | ENST00000479361 | 1229 | PERM1 | GO:0005634(nucleus);GO:0005737(cytoplasm);GO:0006351(transcription, DNA-templated);GO:0006355(regulation of transcription, DNA-templated);GO:0014850(response to muscle activity) | NA | NA | NA | PPARGC1 and ESRR induced regulator, muscle 1 [Source:HGNC Symbol;Acc:HGNC:28208] | 7.95 | -1.49 | -2.64 | -2.53 | down | 1.50 | 0.58 | 0.37 | 1 | up | no |
| chr1 | 1335365 | 1336229 | 865 | ENSG00000107404 | ENST00000632445 | 3193 | DVL1 | GO:0005737(cytoplasm);GO:0016020(membrane);GO:0035556(intracellular signal transduction);GO:0005829(cytosol);GO:0005886(plasma membrane);GO:0005515(protein binding);GO:0007275(multicellular organism development);GO:0016055(Wnt signaling pathway);GO:0031410(cytoplasmic vesicle);GO:0098793(presynapse);GO:0042802(identical protein binding);GO:0006366(transcription from RNA polymerase II promoter);GO:0090090(negative regulation of canonical Wnt signaling pathway);GO:0019899(enzyme binding);GO:0045893(positive regulation of transcription, DNA-templated);GO:0050821(protein stabilization);GO:0060070(canonical Wnt signaling pathway);GO:0099054(presynapse assembly);GO:1905386(positive regulation of protein localization to presynapse);GO:0016328(lateral plasma membrane);GO:0005109(frizzled binding);GO:0019901(protein kinase binding);GO:0001505(regulation of neurotransmitter levels);GO:0006469(negative regulation of protein kinase activity);GO:0007269(neurotransmitter secretion);GO:0007528(neuromuscular junction development);GO:0021915(neural tube development);GO:0030177(positive regulation of Wnt signaling pathway);GO:0032091(negative regulation of protein binding);GO:0032436(positive regulation of proteasomal ubiquitin-dependent protein catabolic process);GO:0034504(protein localization to nucleus);GO:0043113(receptor clustering);GO:0048813(dendrite morphogenesis);GO:0050808(synapse organization);GO:0060071(Wnt signaling pathway, planar cell polarity pathway);GO:0090179(planar cell polarity pathway involved in neural tube closure);GO:1903827(regulation of cellular protein localization);GO:1904886(beta-catenin destruction complex disassembly);GO:0030426(growth cone);GO:0043005(neuron projection);GO:0045202(synapse);GO:0008013(beta-catenin binding);GO:0048365(Rac GTPase binding);GO:0001932(regulation of protein phosphorylation);GO:0001933(negative regulation of protein phosphorylation);GO:0001934(positive regulation of protein phosphorylation);GO:0007409(axonogenesis);GO:0007411(axon guidance);GO:0010976(positive regulation of neuron projection development);GO:0022007(convergent extension involved in neural plate elongation);GO:0031122(cytoplasmic microtubule organization);GO:0035176(social behavior);GO:0035372(protein localization to microtubule);GO:0048668(collateral sprouting);GO:0048675(axon extension);GO:0060029(convergent extension involved in organogenesis);GO:0060134(prepulse inhibition);GO:0060997(dendritic spine morphogenesis);GO:0071340(skeletal muscle acetylcholine-gated channel clustering);GO:0090103(cochlea morphogenesis);GO:0090263(positive regulation of canonical Wnt signaling pathway);GO:2000463(positive regulation of excitatory postsynaptic potential);GO:0005874(microtubule);GO:0014069(postsynaptic density);GO:0015630(microtubule cytoskeleton);GO:0030136(clathrin-coated vesicle);GO:0030424(axon);GO:0030425(dendrite);GO:0043025(neuronal cell body);GO:0043197(dendritic spine);GO:1990909(Wnt signalosome);GO:0005622(intracellular);GO:0005622(intracellular) | 04310(Wnt signaling pathway);04330(Notch signaling pathway);04390(Hippo signaling pathway);04150(mTOR signaling pathway);04550(Signaling pathways regulating pluripotency of stem cells);04916(Melanogenesis);05200(Pathways in cancer);05225(Hepatocellular carcinoma);05226(Gastric cancer);05217(Basal cell carcinoma);05224(Breast cancer);05166(HTLV-I infection);05165(Human papillomavirus infection);05165(Human papillomavirus infection) | K02353 | NA | dishevelled segment polarity protein 1 [Source:HGNC Symbol;Acc:HGNC:3084] | 8.25 | -1.85 | -3.13 | -0.33 | down | 1.15 | 0.21 | 0.71 | 1 | up | no |
Definition of table:
| Term | Description |
|---|---|
| seqnames | The name of the chromosome (e.g. chr3, chrY, chr2 random) or scaffold (e.g. scaffold10671) |
| start | The starting position of the methylation site in the chromosome or scaffold. |
| end | The ending position of the RNA methylation site in the chromosome or scaffold. |
| width | Peak region of methylation |
| geneId | Defines the ID of gene on which the RNA methylation site locates |
| transcriptId | Defines the ID of transcript on which the RNA methylation site locates |
| distanceToTSS | Distrance from Peak site to transcription start site |
| geneName | Defines the name of gene (gene symbol) on which the RNA methylation site locates |
| GO | Gene ontology term and description http://www.geneontology.org/ |
| KEGG | KEGG pathway annotation and description http://www.kegg.jp/kegg/kegg3a.html |
| KO_Entry | KO functional ortholog, KO groups identified by K numbers and, in metabolic maps |
| EC | enzyme |
| Description | Description of gene |
| fold_enrchment | fold enrichment within the peak in the IP sample compared with the input sample. |
| diff.lg.fdr | results from differential methylation analysis, log10(fdr) of the peak as a differential methylation site between the two experimental conditions tested. |
| diff.lg.p | results from differential methylation analysis, log10(pvalue) of the peak as a differential methylation site between the two experimental conditions tested. |
| diff.log2.fc | results from differential methylation analysis, log2(odds ratio) of the peak as a differential methylation site between the two experimental conditions tested. |
| m6A_regulation | up-regulation or down-regulation of methylated genes/transcripts |
| fc | fold change |
| log2(fc) | log2 of fold change value |
| qval | q value or FDR value |
| regulation | up-regulation or down-regulation of genes/transcripts |
| significant | yes or no |
document location: summary/8_diffPeak_differential_expression/*VS*/*_diffPeak_Gene_differential_expression.xlsx
This section is similar with 4.7.5 GO enrichment analysis of differentially expressed genes
document location: summary/8_diffPeak_differential_expression/*VS*/1_GO_Enrichment
This section is similar with 4.7.6 KEGG enrichment analysis of differentially expressed genes
document location: summary/8_diffPeak_differential_expression/*VS*/2_KEGG_Enrichment
2575 West Bellfort Street
Suite 270
Houston, TX
77054 USA
Local (713) 664-7087
Toll Free: 1-888-528-8818
Fax: (713) 664-8181